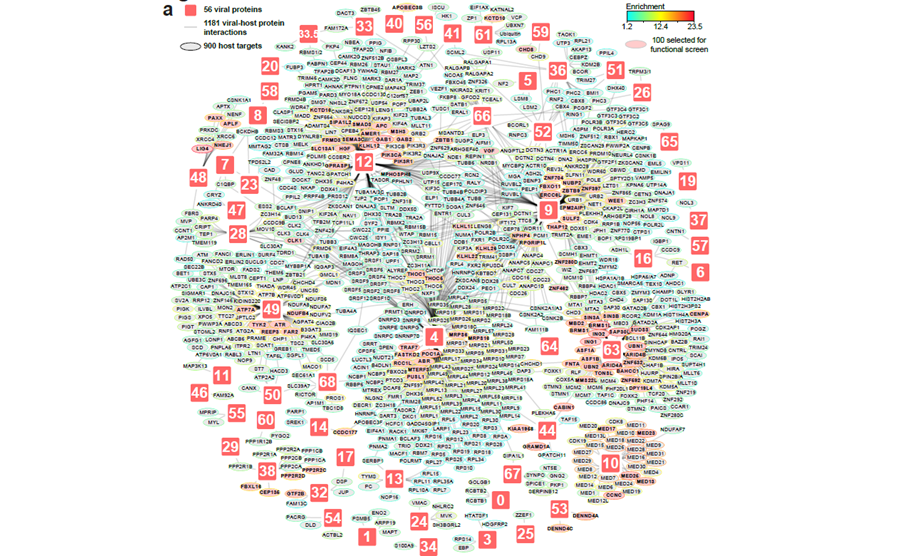
Varicella-zoster virus (VZV), the cause of chickenpox and shingles, is also linked to neurological complications — but how it disrupts human cells at the molecular level has remained unclear. In our latest study published in Nature Microbiology, we systematically mapped how 64 VZV proteins interact with the cellular machinery and induce host protein abundance changes, and thus, individually participate to the proteomic perturbations observed in infected cells. The dataset reveals 900 interactors and thousands of VZV-induced host protein abundance regulations (https://varizonet.innatelab.org). This experimental design allowed comprehensive data integration to spotlighted key molecular strategies mediated by viral proteins. For example, we found a mechanism for the ORF61-mediated IFI16 degradation via the recruitment of host E3 ligase co-factors. Loss-of-function screen of identified host proteins discovered dependency and restriction factors which can be targeted to control VZV infection. Finally, integrating this proteomic VZV–host interface to patient genetic data identified the protein nephrocystin 4 as an important restriction factor, which allowed us to characterise how its variant S862N can participate to increased VZV brain infections.
This work offers a comprehensive herpesvirus–host interface resource, which aids our understanding of the molecular features of disease and data-driven identification of antiviral strategies.
Congratulations to Virginie, Melissa and Susi as well as our alumni Alexey, Darya, Lila, and Antonio, and all the co-authors for this long-lasting and highly collaborative effort!
Last but not least, we thank the patients who participated in this study.
Read more here: Multi-proteomic profiling of the varicella-zoster virus-host interface reveals host susceptibilities to severe infection
Text by Virginie.