A. Stukalov, V. Girault, V. Grass, O. Karayel, V. Bergant, C. Urban, D.A. Haas, Y. Huang, L. Oubraham, A. Wang, M.S. Hamad, A. Piras, F.M. Hansen, M.C. Tanzer, I. Paron, L. Zinzula, T. Enghleitner, M. Reinecke, T.M. Lavacca, R. Ehmann, R. Wölfel, J. Jores, B. Kuster, U. Protzer, R. Rad, J. Ziebuhr, V. Thiel, P. Scaturro, M. Mann, A. Pichlmair
Multilevel Proteomics Reveals Host Perturbations by SARS-CoV-2 and SARS-CoV Journal Article
In: Nature, 2021, ISSN: 0028-0836.
@article{Stukalov2021,
title = {Multilevel Proteomics Reveals Host Perturbations by SARS-CoV-2 and SARS-CoV},
author = {A. Stukalov and V. Girault and V. Grass and O. Karayel and V. Bergant and C. Urban and D.A. Haas and Y. Huang and L. Oubraham and A. Wang and M.S. Hamad and A. Piras and F.M. Hansen and M.C. Tanzer and I. Paron and L. Zinzula and T. Enghleitner and M. Reinecke and T.M. Lavacca and R. Ehmann and R. Wölfel and J. Jores and B. Kuster and U. Protzer and R. Rad and J. Ziebuhr and V. Thiel and P. Scaturro and M. Mann and A. Pichlmair},
doi = {10.1038/s41586-021-03493-4},
issn = {0028-0836},
year = {2021},
date = {2021-04-12},
journal = {Nature},
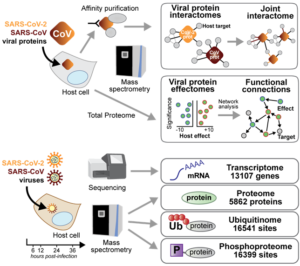
abstract = {The global emergence of SARS-CoV-2 urgently requires an in-depth understanding of molecular functions of viral proteins and their interactions with the host proteome. Several individual omics studies have extended our knowledge of COVID-19 pathophysiology1–10. Integration of such datasets to obtain a holistic view of virus-host interactions and to define the pathogenic properties of SARS-CoV-2 is limited by the heterogeneity of the experimental systems. We therefore conducted a concurrent multi-omics study of SARS-CoV-2 and SARS-CoV. Using state-of-the-art proteomics, we profiled the interactome of both viruses, as well as their influence on transcriptome, proteome, ubiquitinome and phosphoproteome in a lung-derived human cell line. Projecting these data onto the global network of cellular interactions revealed crosstalk between the perturbations taking place upon SARS-CoV-2 and SARS-CoV infections at different layers and identified unique and common molecular mechanisms of these closely related coronaviruses. The TGF-β pathway, known for its involvement in tissue fibrosis, was specifically dysregulated by SARS-CoV-2 ORF8 and autophagy by SARS-CoV-2 ORF3. The extensive dataset (available at https://covinet.innatelab.org) highlights many hotspots that can be targeted by existing drugs and it can guide rational design of virus- and host-directed therapies, which we exemplify by identifying kinase and MMPs inhibitors with potent antiviral effects against SARS-CoV-2.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

P. Hubel, C. Urban, V. Bergant, W.M. Schneider, B. Knauer, A. Stukalov, P. Scaturro, A. Mann, L. Brunotte, H.H. Hoffmann, J.W. Schoggins, M. Schwemmle, M. Mann, C.M. Rice, A. Pichlmair
A Protein-interaction Network of Interferon-stimulated Genes Extends the Innate Immune System Landscape Journal Article
In: Nat. Immunol., 2019, ISSN: 1529-2916.
@article{HubelPichlmair2019_NI,
title = {A Protein-interaction Network of Interferon-stimulated Genes Extends the Innate Immune System Landscape},
author = { P. Hubel and C. Urban and V. Bergant and W.M. Schneider and B. Knauer and A. Stukalov and P. Scaturro and A. Mann and L. Brunotte and H.H. Hoffmann and J.W. Schoggins and M. Schwemmle and M. Mann and C.M. Rice and A. Pichlmair},
url = {https://www.nature.com/articles/s41590-019-0323-3
https://innatelab.virologie.med.tum.de/archives/664},
doi = {10.1038/s41590-019-0323-3},
issn = {1529-2916},
year = {2019},
date = {2019-03-04},
journal = {Nat. Immunol.},
abstract = {Interferon-stimulated genes (ISGs) form the backbone of the innate immune system and are important for limiting intra- and intercellular viral replication and spread. We conducted a mass-spectrometry-based survey to understand the fundamental organization of the innate immune system and to explore the molecular functions of individual ISGs. We identified interactions between 104 ISGs and 1,401 cellular binding partners engaging in 2,734 high-confidence interactions. 90% of these interactions are unreported so far, and our survey therefore illuminates a far wider activity spectrum of ISGs than is currently known. Integration of the resulting ISG-interaction network with published datasets and functional studies allowed us to identify regulators of immunity and processes related to the immune system. Given the extraordinary robustness of the innate immune system, this ISG network may serve as a blueprint for therapeutic targeting of cellular systems to efficiently fight viral infections.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

P. Scaturro, A. Stukalov, D.A. Haas, M. Cortese, K. Draganova, A. Płaszczyca, R. Bartenschlager, M. Götz, A. Pichlmair
An Orthogonal Proteomic Survey Uncovers Novel Zika Virus Host Factors Journal Article
In: Nature, 2018, ISSN: 1476-4687.
@article{Scaturro2018zika,
title = {An Orthogonal Proteomic Survey Uncovers Novel Zika Virus Host Factors},
author = {P. Scaturro and A. Stukalov and D.A. Haas and M. Cortese and K. Draganova and A. Płaszczyca and R. Bartenschlager and M. Götz and A. Pichlmair},
url = {https://www.nature.com/articles/s41586-018-0484-5
https://innatelab.virologie.med.tum.de/archives/458},
doi = {10.1038/s41586-018-0484-5},
issn = {1476-4687},
year = {2018},
date = {2018-09-03},
journal = {Nature},
abstract = {Zika virus (ZIKV) has recently emerged as a global health concern owing to its widespread diffusion and its association with severe neurological symptoms and microcephaly in newborns1. However, the molecular mechanisms that are responsible for the pathogenicity of ZIKV remain largely unknown. Here we use human neural progenitor cells and the neuronal cell line SK-N-BE2 in an integrated proteomics approach to characterize the cellular responses to viral infection at the proteome and phosphoproteome level, and use affinity proteomics to identify cellular targets of ZIKV proteins. Using this approach, we identify 386 ZIKV-interacting proteins, ZIKV-specific and pan-flaviviral activities as well as host factors with known functions in neuronal development, retinal defects and infertility. Moreover, our analysis identified 1,216 phosphorylation sites that are specifically up- or downregulated after ZIKV infection, indicating profound modulation of fundamental signalling pathways such as AKT, MAPK–ERK and ATM–ATR and thereby providing mechanistic insights into the proliferation arrest elicited by ZIKV infection. Functionally, our integrative study identifies ZIKV host-dependency factors and provides a comprehensive framework for a system-level understanding of ZIKV-induced perturbations at the levels of proteins and cellular pathways.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
C. Holze, C. Michaudel, C. Mackowiak, D.A. Haas, C. Benda, P. Hubel, F.L. Pennemann, D. Schnepf, J. Wettmarshausen, M. Braun, D.W. Leung, G.K. Amarasinghe, F. Perocchi, P. Staeheli, B. Ryffel, A. Pichlmair
Oxeiptosis, a ROS-induced Caspase-independent Apoptosis-like Cell-death Pathway Journal Article
In: Nat. Immunol., vol. 19, no. 2, pp. 130-140, 2017.
@article{Holze2017,
title = {Oxeiptosis, a ROS-induced Caspase-independent Apoptosis-like Cell-death Pathway},
author = {C. Holze and C. Michaudel and C. Mackowiak and D.A. Haas and C. Benda and P. Hubel and F.L. Pennemann and D. Schnepf and J. Wettmarshausen and M. Braun and D.W. Leung and G.K. Amarasinghe and F. Perocchi and P. Staeheli and B. Ryffel and A. Pichlmair},
url = {https://www.nature.com/articles/s41590-017-0013-y},
doi = {10.1038/s41590-017-0013-y},
year = {2017},
date = {2017-12-18},
journal = {Nat. Immunol.},
volume = {19},
number = {2},
pages = {130-140},
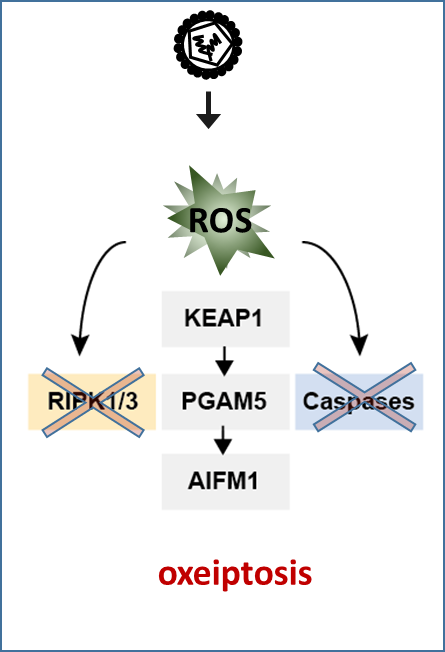
abstract = {Reactive oxygen species (ROS) are generated by virus-infected cells; however, the physiological importance of ROS generated under these conditions is unclear. Here we found that the inflammation and cell death induced by exposure of mice or cells to sources of ROS were not altered in the absence of canonical ROS-sensing pathways or known cell-death pathways. ROS-induced cell-death signaling involved interactions among the cellular ROS sensor and antioxidant factor KEAP1, the phosphatase PGAM5 and the proapoptotic factor AIFM1. Pgam5 –/– mice showed exacerbated lung inflammation and proinflammatory cytokines in an ozone-exposure model. Similarly, challenge with influenza A virus led to increased infiltration of the virus, lymphocytic bronchiolitis and reduced survival of Pgam5 –/– mice. This pathway, which we have called ‘oxeiptosis’, was a ROS-sensitive, caspase independent, non-inflammatory cell-death pathway and was important for protection against inflammation induced by ROS or ROS-generating agents such as viral pathogens.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Alexis Wilhelm, Charlotte Flynn, Evelyn Hammer, Johannes Roessler, Bernhard Haller, Rudolf Napieralski, Moritz Leuthner, Sanja Tosheska, Kèvin Knoops, Anjusha Mathew, Giuliano Ciarimboli, Jan Kranich, Lavinia Flaskamp, Siobhan King, Heidrun Gevensleben, Quirin Emslander, Anna Pastucha, Mathias Reisbeck, Lukas Rief, Holger Bronger, Tobias Dreyer, Andreas R. Bausch, Andreas Pichlmair, Thomas Brocker, Reinhard Zeidler, Wolfgang Hammerschmidt, Melanie Piedavent-Salomom, Carmen López-Iglesias, Gabrielle Schricker, Oliver Haydn, Marion Kiechle, Sabine Grill, Ron Heeren, Percy A. Knolle, Olaf Wilhelm, Bastian Höchst
Two-dimensional analysis of plasma-derived extracellular vesicles to determine the HER2 status in breast cancer patients Journal Article
In: Breast Cancer Res, vol. 27, no. 1, 2025, ISSN: 1465-542X.
@article{Wilhelm2025,
title = {Two-dimensional analysis of plasma-derived extracellular vesicles to determine the HER2 status in breast cancer patients},
author = {Alexis Wilhelm and Charlotte Flynn and Evelyn Hammer and Johannes Roessler and Bernhard Haller and Rudolf Napieralski and Moritz Leuthner and Sanja Tosheska and Kèvin Knoops and Anjusha Mathew and Giuliano Ciarimboli and Jan Kranich and Lavinia Flaskamp and Siobhan King and Heidrun Gevensleben and Quirin Emslander and Anna Pastucha and Mathias Reisbeck and Lukas Rief and Holger Bronger and Tobias Dreyer and Andreas R. Bausch and Andreas Pichlmair and Thomas Brocker and Reinhard Zeidler and Wolfgang Hammerschmidt and Melanie Piedavent-Salomom and Carmen López-Iglesias and Gabrielle Schricker and Oliver Haydn and Marion Kiechle and Sabine Grill and Ron Heeren and Percy A. Knolle and Olaf Wilhelm and Bastian Höchst},
doi = {10.1186/s13058-025-02056-z},
issn = {1465-542X},
year = {2025},
date = {2025-12-00},
journal = {Breast Cancer Res},
volume = {27},
number = {1},
publisher = {Springer Science and Business Media LLC},
abstract = {Abstract
Breast cancer, one of the most common cancers in women, is classified by the expression of hormone receptors and the growth factor receptor HER2, which is important for personalised tumour treatment with HER2-targeted therapies. Tumour biopsies are required for histopathological diagnosis of HER2 expression by breast cancer cells but are subject to sampling error. In this study, we present a method for identifying and analysing cancer-derived EVs from plasma for the detection of HER2 expression in breast cancer without the need for additional processing steps. We detected nano-sized particles through an optimised flow cytometry approach that allows for the identification of HER2-expressing EVs and quantification of their HER2 expression levels. In a clinical study of 115 breast cancer patients, this optimised flow cytometric analysis detected a range of 1.3 to 50 × 103 HER2+ EVs per µl of plasma. The number of HER2+ EVs did not correlate directly with tumour size, grade, or metastasis. However, computational integration of data from the quantification of HER2pos EVs per µl/plasma and their HER2 expression levels on a single EV basis allowed for the reliable identification of HER2 expression levels in tumours. Our results reveal the potential for analysing cancer-derived EVs from plasma for the diagnosis and personalised therapy in breast cancer patients. },
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Franziska Keller, Robert Lorenz Chua, Timo Trefzer, Katharina Jechow, Liane Bauersfeld, Fabian Beier, Sagar, Özlem Sogukpinar, Giuseppe Rusignuolo, Marta Rizzi, Roland Eils, Andreas Pichlmair, Marco Binder, Bertram Bengsch, Christoph Neumann-Haefelin, Volker Lohmann, Tobias Boettler, Christian Conrad, Robert Thimme, Maike Hofmann
Differentiation-associated ISG expression of NK cells in chronic viral infection Journal Article
In: iScience, vol. 28, no. 9, pp. 113216, 2025, ISSN: 2589-0042.
@article{pmid40837234,
title = {Differentiation-associated ISG expression of NK cells in chronic viral infection},
author = {Franziska Keller and Robert Lorenz Chua and Timo Trefzer and Katharina Jechow and Liane Bauersfeld and Fabian Beier and Sagar and Özlem Sogukpinar and Giuseppe Rusignuolo and Marta Rizzi and Roland Eils and Andreas Pichlmair and Marco Binder and Bertram Bengsch and Christoph Neumann-Haefelin and Volker Lohmann and Tobias Boettler and Christian Conrad and Robert Thimme and Maike Hofmann},
doi = {10.1016/j.isci.2025.113216},
issn = {2589-0042},
year = {2025},
date = {2025-09-01},
journal = {iScience},
volume = {28},
number = {9},
pages = {113216},
abstract = {Natural killer (NK) cell responses are modulated by type-I interferons (IFNs) in viral infection. Chronic hepatitis C virus (HCV) infection, marked by robust IFN signatures, shows NK cells with reduced cytokine release but heightened cytotoxicity. Comparable alterations occur in chronic hepatitis B virus (HBV) infection even without a pronounced IFN milieu, implying additional regulatory layers. We analyzed NK cells from healthy donors and patients with chronic HBV or HCV and found conserved expression patterns of interferon-stimulated genes (ISGs) such as , , and that correlated with NK cell differentiation state. These genes are governed by fate-determining transcription factors, including ETS1, FLI1, and Eomes, and appear to be constitutively expressed rather than driven by persistent IFN exposure. Network analysis suggested that NK cell ISGs participate not only in antiviral defense but also in processes such as transport and metabolism, underscoring their role in shaping NK responses during health and chronic viral infection.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Virginie Girault, Alexey Stukalov, Madalina Elena Carter-Timofte, Jonny Hertzog, Melissa Verin, Katharina Austen, Darya A. Haas, Lila Oubraham, Antonio Piras, Susanne Maidl, Rupert Öllinger, Roland Rad, Ulrike Protzer, Benedikt B. Kaufer, Robert J. Lebbink, Jan Rehwinkel, Trine H. Mogensen, Andreas Pichlmair
Multi-proteomic profiling of the varicella-zoster virus–host interface reveals host susceptibilities to severe infection Journal Article
In: Nat Microbiol, vol. 10, no. 8, pp. 2048–2072, 2025, ISSN: 2058-5276.
@article{Girault2025,
title = {Multi-proteomic profiling of the varicella-zoster virus–host interface reveals host susceptibilities to severe infection},
author = {Virginie Girault and Alexey Stukalov and Madalina Elena Carter-Timofte and Jonny Hertzog and Melissa Verin and Katharina Austen and Darya A. Haas and Lila Oubraham and Antonio Piras and Susanne Maidl and Rupert Öllinger and Roland Rad and Ulrike Protzer and Benedikt B. Kaufer and Robert J. Lebbink and Jan Rehwinkel and Trine H. Mogensen and Andreas Pichlmair},
doi = {10.1038/s41564-025-02068-7},
issn = {2058-5276},
year = {2025},
date = {2025-08-00},
journal = {Nat Microbiol},
volume = {10},
number = {8},
pages = {2048--2072},
publisher = {Springer Science and Business Media LLC},
abstract = {Abstract
Varicella-zoster virus (VZV) infects most humans and causes chickenpox, shingles and central nervous system pathologies. The molecular basis for these phenotypes remains elusive. Here we conducted a multi-proteomic survey on 64 individual VZV proteins and infection-induced perturbations in a neuronal cell line, identifying 900 interactors and 3,618 regulated host proteins. Data integration suggested molecular functions of viral proteins, such as a mechanism for the ORF61-mediated IFI16 degradation via the recruitment of E3 ligase co-factors. Moreover, we identified proviral host factors (MPP8 and ZNF280D) as potential targets to limit infection. Integration of exome sequencing analysis from patients with VZV-associated central nervous system pathologies identified nephrocystin 4 as a viral restriction factor, and its S862N variant, which showed reduced activity and decreased binding to the regulatory proteins 14-3-3. Collectively, our study provides a comprehensive herpesvirus–host interface resource, which aids our understanding of disease-associated molecular perturbations and data-driven identification of antiviral treatment options. },
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Susanne Klute, Rayhane Nchioua, Arne Cordsmeier, Jyoti Vishwakarma, Lennart Koepke, Hala Alshammary, Christoph Jung, Maximilian Hirschenberger, Helene Hoenigsperger, Jana-Romana Fischer, Rinu Sivarajan, Fabian Zech, Steffen Stenger, Ruth Serra-Moreno, Ana Silvia Gonzalez-Reiche, Emilia Mia Sordillo, Harm van Bakel, Viviana Simon, Frank Kirchhoff, Timo Jacob, Dorota Kmiec, Andreas Pichlmair, Armin Ensser, Konstantin Maria Johannes Sparrer
Mutation T9I in Envelope confers autophagy resistance to SARS-CoV-2 Omicron Journal Article
In: iScience, vol. 28, no. 7, 2025, ISSN: 2589-0042.
@article{Klute2025,
title = {Mutation T9I in Envelope confers autophagy resistance to SARS-CoV-2 Omicron},
author = {Susanne Klute and Rayhane Nchioua and Arne Cordsmeier and Jyoti Vishwakarma and Lennart Koepke and Hala Alshammary and Christoph Jung and Maximilian Hirschenberger and Helene Hoenigsperger and Jana-Romana Fischer and Rinu Sivarajan and Fabian Zech and Steffen Stenger and Ruth Serra-Moreno and Ana Silvia Gonzalez-Reiche and Emilia Mia Sordillo and Harm van Bakel and Viviana Simon and Frank Kirchhoff and Timo Jacob and Dorota Kmiec and Andreas Pichlmair and Armin Ensser and Konstantin Maria Johannes Sparrer},
doi = {10.1016/j.isci.2025.112974},
issn = {2589-0042},
year = {2025},
date = {2025-07-00},
journal = {iScience},
volume = {28},
number = {7},
publisher = {Elsevier BV},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Veronika Merold, Indra Bekere, Stefanie Kretschmer, Adrian F. Schnell, Dorota Kmiec, Rinu Sivarajan, Katja Lammens, Rou Liu, Julia Mergner, Julia Teppert, Maximilian Hirschenberger, Alexander Henrici, Sarah Hammes, Kathrin Buder, Marcus Weitz, Karl Hackmann, Lars M. Koenig, Andreas Pichlmair, Nadine Schwierz, Konstantin M. J. Sparrer, Min Ae Lee-Kirsch, Carina C. Oliveira Mann
Structural basis for OAS2 regulation and its antiviral function Journal Article
In: Molecular Cell, vol. 85, no. 11, pp. 2176-2193.e13, 2025, ISSN: 1097-2765.
@article{MEROLD20252176,
title = {Structural basis for OAS2 regulation and its antiviral function},
author = {Veronika Merold and Indra Bekere and Stefanie Kretschmer and Adrian F. Schnell and Dorota Kmiec and Rinu Sivarajan and Katja Lammens and Rou Liu and Julia Mergner and Julia Teppert and Maximilian Hirschenberger and Alexander Henrici and Sarah Hammes and Kathrin Buder and Marcus Weitz and Karl Hackmann and Lars M. Koenig and Andreas Pichlmair and Nadine Schwierz and Konstantin M. J. Sparrer and Min Ae Lee-Kirsch and Carina C. Oliveira Mann},
url = {https://www.sciencedirect.com/science/article/pii/S109727652500406X},
doi = {https://doi.org/10.1016/j.molcel.2025.05.001},
issn = {1097-2765},
year = {2025},
date = {2025-01-01},
journal = {Molecular Cell},
volume = {85},
number = {11},
pages = {2176-2193.e13},
abstract = {Summary
Oligoadenylate synthetase (OAS) proteins are immune sensors for double-stranded RNA and are critical for restricting viruses. OAS2 comprises two OAS domains, only one of which can synthesize 2′–5′-oligoadenylates for RNase L activation. Existing structures of OAS1 provide a model for enzyme activation, but they do not explain how multiple OAS domains discriminate RNA length. Here, we discover that human OAS2 exists in an auto-inhibited state as a zinc-mediated dimer and present a mechanism for RNA length discrimination: the catalytically deficient domain acts as a molecular ruler that prevents autoreactivity to short RNAs. We demonstrate that dimerization and myristoylation localize OAS2 to Golgi membranes and that this is required for OAS2 activation and the restriction of viruses that exploit the endomembrane system for replication, e.g., coronaviruses. Finally, our results highlight the non-redundant role of OAS proteins and emphasize the clinical relevance of OAS2 by identifying a patient with a loss-of-function mutation associated with autoimmune disease.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Stefan Enssle, Anna Sax, Peter May, Nadia El Khawanky, Nardine Soliman, Markus Perl, Julius C. Enssle, Karsten Krey, Jürgen Ruland, Andreas Pichlmair, Florian Bassermann, Hendrik Poeck, Simon Heidegger
In: OncoImmunology, vol. 14, no. 1, pp. 2504244, 2025, (PMID: 40366863).
@article{Enssle31122025,
title = {Gasdermin E links tumor cell-intrinsic nucleic acid signaling to proinflammatory cell death for successful checkpoint inhibitor cancer immunotherapy},
author = {Stefan Enssle and Anna Sax and Peter May and Nadia El Khawanky and Nardine Soliman and Markus Perl and Julius C. Enssle and Karsten Krey and Jürgen Ruland and Andreas Pichlmair and Florian Bassermann and Hendrik Poeck and Simon Heidegger},
url = {https://doi.org/10.1080/2162402X.2025.2504244},
doi = {10.1080/2162402X.2025.2504244},
year = {2025},
date = {2025-01-01},
journal = {OncoImmunology},
volume = {14},
number = {1},
pages = {2504244},
publisher = {Taylor & Francis},
note = {PMID: 40366863},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Teng-Feng Li, Paul Rothhaar, Arthur Lang, Oliver Grünvogel, Ombretta Colasanti, Santa Mariela Olivera Ugarte, Jannik Traut, Antonio Piras, Nelson Acosta-Rivero, Vladimir Gonçalves Magalhães, Emely Springer, Andreas Betz, Hao-En Huang, Jeongbin Park, Ruiyue Qiu, Gnimah Eva Gnouamozi, Ann-Kathrin Mehnert, Viet Loan Dao Thi, Stephan Urban, Martina Muckenthaler, Matthias Schlesner, Dirk Wohlleber, Marco Binder, Ralf Bartenschlager, Andreas Pichlmair, Volker Lohmann
RBM39 shapes innate immunity by controlling the expression of key factors of the interferon response Journal Article
In: Frontiers in Immunology, vol. Volume 16 - 2025, 2025, ISSN: 1664-3224.
@article{10.3389/fimmu.2025.1568056,
title = {RBM39 shapes innate immunity by controlling the expression of key factors of the interferon response},
author = {Teng-Feng Li and Paul Rothhaar and Arthur Lang and Oliver Grünvogel and Ombretta Colasanti and Santa Mariela Olivera Ugarte and Jannik Traut and Antonio Piras and Nelson Acosta-Rivero and Vladimir Gonçalves Magalhães and Emely Springer and Andreas Betz and Hao-En Huang and Jeongbin Park and Ruiyue Qiu and Gnimah Eva Gnouamozi and Ann-Kathrin Mehnert and Viet Loan Dao Thi and Stephan Urban and Martina Muckenthaler and Matthias Schlesner and Dirk Wohlleber and Marco Binder and Ralf Bartenschlager and Andreas Pichlmair and Volker Lohmann},
url = {https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2025.1568056},
doi = {10.3389/fimmu.2025.1568056},
issn = {1664-3224},
year = {2025},
date = {2025-01-01},
journal = {Frontiers in Immunology},
volume = {Volume 16 - 2025},
abstract = {Background and aimsThe contribution of innate immunity to clearance of viral infections of the liver, in particular sensing via Toll-like receptor 3 (TLR3), is incompletely understood. We aimed to identify the factors contributing to the TLR3 response in hepatocytes via CRISPR/Cas9 screening.MethodsA genome-wide CRISPR/Cas9 screen on the TLR3 pathway was performed in two liver-derived cell lines, followed by siRNA knockdown validation. SiRNA knockdown and indisulam treatment were used to study the role of RNA-binding motif protein 39 (RBM39) in innate immunity upon poly(I:C) or cytokine treatment and viral infections. Transcriptome, proteome, and alternative splicing were studied via RNA sequencing and mass spectrometry upon depletion of RBM39.ResultsOur CRISPR/Cas9 screen identified RBM39, which is highly expressed in hepatocytes, as an important regulator of the TLR3 pathway. Knockdown of RBM39 or treatment with indisulam, an aryl sulfonamide drug targeting RBM39 for proteasomal degradation, strongly reduced the induction of interferon-stimulated genes (ISGs) in response to double-stranded RNA (dsRNA) or viral infections. RNA sequencing (seq) and mass spectrometry identified that transcription and/or splicing of the key pathway components IRF3, RIG-I, and MDA5 were affected by RBM39 depletion, along with multiple other cellular processes identified previously. RBM39 knockdown further restrained type I and type III IFN pathways by reducing the expression of individual receptor subunits and STAT1/2. The function of RBM39 was furthermore not restricted to hepatocytes.ConclusionWe identified RBM39 as a regulatory factor of cell intrinsic innate immune signaling. Depletion of RBM39 impaired TLR3, RIG-I/MDA5, and IFN responses by affecting the basal expression of key pathway components.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Charlotte Rothfuß, Tobias Baumann, Sainitin Donakonda, Bettina Brauchle, Anetta Marcinek, Christian Urban, Julia Mergner, Anna-Marie Pedde, Anna Hirschberger, Christina Krupka, Anne-Sophie Neumann, Gerulf Hänel, Camilla Merten, Rupert Öllinger, Judith S Hecker, Tanja Bauer, Christian Rudolf Schmid, Katharina S Götze, Jennifer Altomonte, Veit L Bücklein, Roland Jacobs, Roland Rad, Corinna Dawid, Luca Simeoni, Burkhart Schraven, Andreas Pichlmair, Marion Subklewe, Percy Alexander Knolle, Jan P Böttcher, Bastian Höchst
Two-layered immune escape in AML is overcome by Fcγ receptor activation and inhibition of PGE2 signaling in NK cells Journal Article
In: Blood, 2025, ISSN: 1528-0020.
@article{pmid39840945,
title = {Two-layered immune escape in AML is overcome by Fcγ receptor activation and inhibition of PGE2 signaling in NK cells},
author = {Charlotte Rothfuß and Tobias Baumann and Sainitin Donakonda and Bettina Brauchle and Anetta Marcinek and Christian Urban and Julia Mergner and Anna-Marie Pedde and Anna Hirschberger and Christina Krupka and Anne-Sophie Neumann and Gerulf Hänel and Camilla Merten and Rupert Öllinger and Judith S Hecker and Tanja Bauer and Christian Rudolf Schmid and Katharina S Götze and Jennifer Altomonte and Veit L Bücklein and Roland Jacobs and Roland Rad and Corinna Dawid and Luca Simeoni and Burkhart Schraven and Andreas Pichlmair and Marion Subklewe and Percy Alexander Knolle and Jan P Böttcher and Bastian Höchst},
doi = {10.1182/blood.2024025706},
issn = {1528-0020},
year = {2025},
date = {2025-01-01},
journal = {Blood},
abstract = {Loss of anticancer NK cell function in AML patients is associated with fatal disease progression and remains poorly understood. Here, we demonstrate that AML-blasts isolated from patients rapidly inhibit NK cell function and escape NK cell-mediated killing. Transcriptome analysis of NK cells exposed to AML-blasts revealed increased CREM expression and transcriptional activity, indicating enhanced cAMP signalling, confirmed by uniform production of the cAMP-inducing prostanoid PGE2 by all AML-blast isolates from patients. Phosphoproteome analysis disclosed that PGE2 induced a blockade of LCK-ERK signalling that is crucial for NK cell activation, indicating a two-layered escape of AML-blasts with low expression of NK cell-activating ligands and inhibition of NK cell signalling. To evaluate the therapeutic potential to target PGE2 inhibition, we combined Fcg-receptor-mediated activation with the prevention of inhibitory PGE2-signalling. This rescued NK cell function and restored the killing of AML-blasts. Thus, we identify the PGE2-LCK signalling axis as the key barrier for NK cell activation in two-layered immune escape of AML-blasts that can be targeted for immune therapy to reconstitute anti-cancer NK cell immunity in AML patients.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Wesley Freppel, Viviana Andrea Barragan Torres, Olus Uyar, Anaïs Anton, Zaynab Nouhi, Mathilde Broquière, Clément Mazeaud, Aïssatou Aïcha Sow, Alexanne Léveillé, Claudia Gilbert, Nicolas Tremblay, Jonathan Eintrez Owen, Cheyanne L Bemis, Xavier Laulhé, Alain Lamarre, Christopher J Neufeldt, Ian Gaël Rodrigue-Gervais, Andreas Pichlmair, Denis Girard, Pietro Scaturro, Laura Hulea, Laurent Chatel-Chaix
Dengue virus and Zika virus alter endoplasmic reticulum-mitochondria contact sites to regulate respiration and apoptosis Journal Article
In: iScience, vol. 28, no. 1, pp. 111599, 2025, ISSN: 2589-0042.
@article{pmid39834870,
title = {Dengue virus and Zika virus alter endoplasmic reticulum-mitochondria contact sites to regulate respiration and apoptosis},
author = {Wesley Freppel and Viviana Andrea Barragan Torres and Olus Uyar and Anaïs Anton and Zaynab Nouhi and Mathilde Broquière and Clément Mazeaud and Aïssatou Aïcha Sow and Alexanne Léveillé and Claudia Gilbert and Nicolas Tremblay and Jonathan Eintrez Owen and Cheyanne L Bemis and Xavier Laulhé and Alain Lamarre and Christopher J Neufeldt and Ian Gaël Rodrigue-Gervais and Andreas Pichlmair and Denis Girard and Pietro Scaturro and Laura Hulea and Laurent Chatel-Chaix},
doi = {10.1016/j.isci.2024.111599},
issn = {2589-0042},
year = {2025},
date = {2025-01-01},
journal = {iScience},
volume = {28},
number = {1},
pages = {111599},
abstract = {During infection, dengue virus (DENV) and Zika virus (ZIKV), two (ortho)flaviviruses of public health concern worldwide, induce alterations of mitochondria morphology to favor viral replication, suggesting a viral co-opting of mitochondria functions. Here, we performed an extensive transmission electron microscopy-based quantitative analysis to demonstrate that both DENV and ZIKV alter endoplasmic reticulum-mitochondria contact sites (ERMC). This correlated at the molecular level with an impairment of ERMC tethering protein complexes located at the surface of both organelles. Furthermore, virus infection modulated the mitochondrial oxygen consumption rate. Consistently, metabolomic and mitoproteomic analyses revealed a decrease in the abundance of several metabolites of the Krebs cycle and changes in the stoichiometry of the electron transport chain. Most importantly, ERMC destabilization by protein knockdown increased virus replication while dampening ZIKV-induced apoptosis. Overall, our results support the notion that flaviviruses hijack ERMCs to generate a cytoplasmic environment beneficial for sustained and efficient replication.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Chunkyu Ko, Cho-Chin Cheng, Daniele Mistretta, Shubhankar Ambike, Julia Sacherl, Stoyan Velkov, Bo-Hung Liao, Romina Bester, Merve Gültan, Olga Polezhaeva, Alexander Herrmann, Constanze A Jakwerth, Carsten B Schmidt-Weber, Joachim J Bugert, Roman Wölfel, Vincent Grass, Sandra Essbauer, Daniel Schnepf, Oliver T Keppler, Florian W R Vondran, Andreas Pichlmair, Carolin Mogler, Gregor Ebert, Ulrike Protzer
SARS-CoV-2 Productively Infects Human Hepatocytes and Induces Cell Death Journal Article
In: J Med Virol, vol. 97, no. 1, pp. e70156, 2025, ISSN: 1096-9071.
@article{pmid39760326,
title = {SARS-CoV-2 Productively Infects Human Hepatocytes and Induces Cell Death},
author = {Chunkyu Ko and Cho-Chin Cheng and Daniele Mistretta and Shubhankar Ambike and Julia Sacherl and Stoyan Velkov and Bo-Hung Liao and Romina Bester and Merve Gültan and Olga Polezhaeva and Alexander Herrmann and Constanze A Jakwerth and Carsten B Schmidt-Weber and Joachim J Bugert and Roman Wölfel and Vincent Grass and Sandra Essbauer and Daniel Schnepf and Oliver T Keppler and Florian W R Vondran and Andreas Pichlmair and Carolin Mogler and Gregor Ebert and Ulrike Protzer},
doi = {10.1002/jmv.70156},
issn = {1096-9071},
year = {2025},
date = {2025-01-01},
journal = {J Med Virol},
volume = {97},
number = {1},
pages = {e70156},
abstract = {SARS-CoV-2 infection is accompanied by elevated liver enzymes, and patients with pre-existing liver conditions experience more severe disease. While it was known that SARS-CoV-2 infects human hepatocytes, our study determines the mechanism of infection, demonstrates viral replication and spread, and highlights direct hepatocyte damage. Viral replication was readily detectable upon infection of primary human hepatocytes and hepatoma cells with the ancestral SARS-CoV-2, Delta, and Omicron variants. Hepatocytes express the SARS-CoV-2 receptor ACE2 and the host cell protease TMPRSS2, and knocking down ACE2 and TMPRSS2 impaired SARS-CoV-2 infection. Progeny viruses released from infected hepatocytes showed the typical coronavirus morphology by electron microscopy and proved infectious when transferred to fresh cells, indicating that hepatocytes can contribute to virus spread. Importantly, SARS-CoV-2 infection rapidly induced hepatocyte death in a replication-dependent fashion, with the Omicron variant showing faster onset but less extensive cell death. C57BL/6 wild-type mice infected with a mouse-adapted SARS-CoV-2 strain showed high levels of viral RNA in liver and lung tissues. ALT peaked when viral RNA was cleared from the liver. Liver histology revealed profound tissue damage and immune cell infiltration, indicating that direct cytopathic effects of SARS-CoV-2 and immune-mediated killing of infected hepatocytes contribute to liver pathology.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Alexander Kirchhoff, Anna-Maria Herzner, Christian Urban, Antonio Piras, Robert Düster, Julia Mahlberg, Agathe Grünewald, Thais M Schlee-Guimarães, Katrin Ciupka, Petro Leka, Robert J Bootz, Christina Wallerath, Charlotte Hunkler, Ann Kristin de Regt, Beate M Kümmerer, Maria Hønholt Christensen, Florian I Schmidt, Min Ae Lee-Kirsch, Claudia Günther, Hiroki Kato, Eva Bartok, Gunther Hartmann, Matthias Geyer, Andreas Pichlmair, Martin Schlee
RNA-binding proteins hnRNPM and ELAVL1 promote type-I interferon induction downstream of the nucleic acid sensors cGAS and RIG-I Journal Article
In: EMBO J, 2024, ISSN: 1460-2075.
@article{pmid39707025,
title = {RNA-binding proteins hnRNPM and ELAVL1 promote type-I interferon induction downstream of the nucleic acid sensors cGAS and RIG-I},
author = {Alexander Kirchhoff and Anna-Maria Herzner and Christian Urban and Antonio Piras and Robert Düster and Julia Mahlberg and Agathe Grünewald and Thais M Schlee-Guimarães and Katrin Ciupka and Petro Leka and Robert J Bootz and Christina Wallerath and Charlotte Hunkler and Ann Kristin de Regt and Beate M Kümmerer and Maria Hønholt Christensen and Florian I Schmidt and Min Ae Lee-Kirsch and Claudia Günther and Hiroki Kato and Eva Bartok and Gunther Hartmann and Matthias Geyer and Andreas Pichlmair and Martin Schlee},
doi = {10.1038/s44318-024-00331-x},
issn = {1460-2075},
year = {2024},
date = {2024-12-01},
journal = {EMBO J},
abstract = {The cytosolic nucleic acid sensors RIG-I and cGAS induce type-I interferon (IFN)-mediated immune responses to RNA and DNA viruses, respectively. So far no connection between the two cytosolic pathways upstream of IKK-like kinase activation has been investigated. Here, we identify heterogeneous nuclear ribonucleoprotein M (hnRNPM) as a positive regulator of IRF3 phosphorylation and type-I IFN induction downstream of both cGAS and RIG-I. Combining interactome analysis with genome editing, we further uncover the RNA-binding protein ELAV-like protein 1 (ELAVL1; also known as human antigen R, HuR) as an hnRNPM interactor. Depletion of hnRNPM or ELAVL1 impairs type-I IFN induction by herpes simplex virus 1 or Sendai virus. In addition, we show that hnRNPM and ELAVL1 interact with TANK-binding kinase 1, IκB kinase ε, IκB kinase β, and NF-κB p65. Our confocal microscopy experiments demonstrate cytosolic and perinuclear interactions between hnRNPM, ELAVL1, and TBK1. Furthermore, pharmacological inhibition of ELAVL1 strongly reduces cytokine release from type-I interferonopathy patient fibroblasts. The RNA-binding proteins hnRNPM and ELAVL1 are the first non-redundant regulators to bridge the cGAS/STING and RIG-I/MAVS pathways. Overall, our study characterizes the hnRNPM-ELAVL1 complex as a novel system promoting antiviral defense, pointing to a potential therapeutic target to reduce auto-inflammation in patients with type-I interferonopathies.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Søren R Paludan, Thomas Pradeu, Andreas Pichlmair, K Brad Wray, Jacob Giehm Mikkelsen, David Olagnier, Trine H Mogensen
Early host defense against virus infections Journal Article
In: Cell Rep, vol. 43, no. 12, pp. 115070, 2024, ISSN: 2211-1247.
@article{pmid39675007,
title = {Early host defense against virus infections},
author = {Søren R Paludan and Thomas Pradeu and Andreas Pichlmair and K Brad Wray and Jacob Giehm Mikkelsen and David Olagnier and Trine H Mogensen},
doi = {10.1016/j.celrep.2024.115070},
issn = {2211-1247},
year = {2024},
date = {2024-12-01},
journal = {Cell Rep},
volume = {43},
number = {12},
pages = {115070},
abstract = {Early host defense eliminates many viruses before infections are established while clearing others so they remain subclinical or cause only mild disease. The field of immunology has been shaped by broad concepts, including the pattern recognition theory that currently dominates innate immunology. Focusing on early host responses to virus infections, we analyze the literature to build a working hypothesis for the principles that govern the early line of cellular antiviral defense. Aiming to ultimately arrive at a criteria-based theory with strong explanatory power, we propose that both controlling infection and limiting inflammation are key drivers for the early cellular antiviral response. This response, which we suggest is exerted by a set of "microbe- and inflammation-restricting mechanisms," directly restrict viral replication while also counteracting inflammation. Exploring the mechanisms and physiological importance of the early layer of cellular antiviral defense may open further lines of research in immunology.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Christiane Guder, Soraya Heinrich, Vanadin Seifert-Klauss, Marion Kiechle, Lisa Bauer, Rupert Öllinger, Andreas Pichlmair, Marie-Nicole Theodoraki, Veena Ramesh, Ali Bashiri Dezfouli, Barbara Wollenberg, Alan Graham Pockley, Gabriele Multhoff
Extracellular Hsp70 and Circulating Endometriotic Cells as Novel Biomarkers for Endometriosis Journal Article
In: Int J Mol Sci, vol. 25, no. 21, 2024, ISSN: 1422-0067.
@article{pmid39519195,
title = {Extracellular Hsp70 and Circulating Endometriotic Cells as Novel Biomarkers for Endometriosis},
author = {Christiane Guder and Soraya Heinrich and Vanadin Seifert-Klauss and Marion Kiechle and Lisa Bauer and Rupert Öllinger and Andreas Pichlmair and Marie-Nicole Theodoraki and Veena Ramesh and Ali Bashiri Dezfouli and Barbara Wollenberg and Alan Graham Pockley and Gabriele Multhoff},
doi = {10.3390/ijms252111643},
issn = {1422-0067},
year = {2024},
date = {2024-10-01},
journal = {Int J Mol Sci},
volume = {25},
number = {21},
abstract = {Stress-inducible heat shock protein 70 (Hsp70), which functions as a molecular chaperone and is frequently overexpressed in different cancer cell types, is present on the cell surface of tumor cells and is actively released into the circulation in free and extracellular lipid vesicle-associated forms. Since the exact pathomechanism of endometriosis has not yet been elucidated (although it has been associated with the development of endometrial and ovarian cancer), we asked whether extracellular Hsp70 and circulating endometriotic cells (CECs) reflect the presence and development of endometriosis. Therefore, circulating levels of free and lipid microvesicle-associated Hsp70 were measured using the Hsp70-exo ELISA, and the presence of circulating CECs in the peripheral blood of patients with endometriosis was determined using membrane Hsp70 (mHsp70) and EpCAM monoclonal antibody (mAb)-based bead isolation approaches. Isolated CECs were further characterized by immunofluorescence using reagents directed against cytokeratin (epithelial marker), CD45 (leukocyte marker), CD105/CD44 (mesenchymal stemness markers) and by comparative RNA analysis. Similar to the situation in patients with cancer, the levels of circulating Hsp70 were elevated in the blood of patients with histologically proven endometriosis compared to a healthy control cohort, with significantly elevated Hsp70 levels in endometriosis patients with lesions outside the uterine cavity. Moreover, CECs could be isolated using the cmHsp70.1 mAb-based, and to a lesser extent EpCAM mAb-based, bead approach in all patients with endometriosis, with the highest counts obtained using the mHsp70-targeting procedure in patients with extra-uterine involvement. The longevity in cell culture and the expression of the cytokeratins CD105 and CD44, together with differentially expressed genes related to epithelial-to-mesenchymal transition (EMT), revealed similarities between mHsp70-expressing CECs and circulating tumor cells (CTCs) and suggest a mesenchymal stem cell origin. These findings support the involvement of mHsp70-positive stem cell-like cells in the development of endometriotic lesions. In summary, elevated levels of Hsp70 and CECs in the circulation could serve as liquid biopsy markers for endometriosis with extra-uterine involvement and help to elucidate the underlying pathomechanism of the disease.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Yiqi Huang, Christian Urban, Philipp Hubel, Alexey Stukalov, Andreas Pichlmair
Protein turnover regulation is critical for influenza A virus infection Journal Article
In: Cell Syst, vol. 15, no. 10, pp. 911–929.e8, 2024, ISSN: 2405-4720.
@article{pmid39368468,
title = {Protein turnover regulation is critical for influenza A virus infection},
author = {Yiqi Huang and Christian Urban and Philipp Hubel and Alexey Stukalov and Andreas Pichlmair},
doi = {10.1016/j.cels.2024.09.004},
issn = {2405-4720},
year = {2024},
date = {2024-10-01},
journal = {Cell Syst},
volume = {15},
number = {10},
pages = {911--929.e8},
abstract = {The abundance of a protein is defined by its continuous synthesis and degradation, a process known as protein turnover. Here, we systematically profiled the turnover of proteins in influenza A virus (IAV)-infected cells using a pulse-chase stable isotope labeling by amino acids in cell culture (SILAC)-based approach combined with downstream statistical modeling. We identified 1,798 virus-affected proteins with turnover changes (tVAPs) out of 7,739 detected proteins (data available at pulsechase.innatelab.org). In particular, the affected proteins were involved in RNA transcription, splicing and nuclear transport, protein translation and stability, and energy metabolism. Many tVAPs appeared to be known IAV-interacting proteins that regulate virus propagation, such as KPNA6, PPP6C, and POLR2A. Notably, our analysis identified additional IAV host and restriction factors, such as the splicing factor GPKOW, that exhibit significant turnover rate changes while their total abundance is minimally affected. Overall, we show that protein turnover is a critical factor both for virus replication and antiviral defense.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Anna Macht, Yiqi Huang, Line S Reinert, Vincent Grass, Kristin Lohmer, Elke Tatjana Aristizabal Prada, Eveline Babel, Alexandra Semmler, Wen Zhang, Andrea Wegner, Eva Lichtenegger-Hartl, Sonja Haas, Günther Hasenpusch, Steffen Meyer, Søren R Paludan, Andreas Pichlmair, Carsten Rudolph, Thomas Langenickel
Mucosal IFNλ1 mRNA-based immunomodulation effectively reduces SARS-CoV-2 induced mortality in mice Journal Article
In: EMBO Rep, vol. 25, no. 9, pp. 3777–3788, 2024, ISSN: 1469-3178.
@article{pmid39060455,
title = {Mucosal IFNλ1 mRNA-based immunomodulation effectively reduces SARS-CoV-2 induced mortality in mice},
author = {Anna Macht and Yiqi Huang and Line S Reinert and Vincent Grass and Kristin Lohmer and Elke Tatjana Aristizabal Prada and Eveline Babel and Alexandra Semmler and Wen Zhang and Andrea Wegner and Eva Lichtenegger-Hartl and Sonja Haas and Günther Hasenpusch and Steffen Meyer and Søren R Paludan and Andreas Pichlmair and Carsten Rudolph and Thomas Langenickel},
doi = {10.1038/s44319-024-00216-4},
issn = {1469-3178},
year = {2024},
date = {2024-09-01},
journal = {EMBO Rep},
volume = {25},
number = {9},
pages = {3777--3788},
abstract = {RNA vaccines elicit protective immunity against SARS-CoV-2, but the use of mRNA as an antiviral immunotherapeutic is unexplored. Here, we investigate the activity of lipidoid nanoparticle (LNP)-formulated mRNA encoding human IFNλ1 (ETH47), which is a critical driver of innate immunity at mucosal surfaces protecting from viral infections. IFNλ1 mRNA administration promotes dose-dependent protein translation, induction of interferon-stimulated genes without relevant signs of unspecific immune stimulation, and dose-dependent inhibition of SARS-CoV-2 replication in vitro. Pulmonary administration of IFNλ1 mRNA in mice results in a potent reduction of virus load, virus-induced body weight loss and significantly increased survival. These data support the development of inhaled administration of IFNλ1 mRNA as a potential prophylactic option for individuals exposed to SARS-CoV-2 or at risk suffering from COVID-19. Based on the broad antiviral activity of IFNλ1 regardless of virus or variant, this approach might also be utilized for other respiratory viral infections or pandemic preparedness.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Mariya Al Hamrashdi, Carla Sanchez Perez, Darya A Haas, Jyoti Vishwakarma, Andreas Pichlmair, Andrew G Bowie, Gareth Brady
Molluscum contagiosum virus protein MC089 inhibits interferon regulatory factor 3 activation Journal Article
In: J Gen Virol, vol. 105, no. 8, 2024, ISSN: 1465-2099.
@article{pmid39167082,
title = {Molluscum contagiosum virus protein MC089 inhibits interferon regulatory factor 3 activation},
author = {Mariya Al Hamrashdi and Carla Sanchez Perez and Darya A Haas and Jyoti Vishwakarma and Andreas Pichlmair and Andrew G Bowie and Gareth Brady},
doi = {10.1099/jgv.0.002015},
issn = {1465-2099},
year = {2024},
date = {2024-08-01},
journal = {J Gen Virol},
volume = {105},
number = {8},
abstract = {Molluscum contagiosum virus (MCV) is a human-specific poxvirus that causes a highly common but mild infection characterized by distinctive and persistent papular skin lesions. These lesions can persist for long periods without an effective clearance response from the host. MCV, like all poxviruses, encodes multiple known immunosuppressive proteins which target innate immune signalling pathways involved in viral nucleic acid sensing, interferon production and inflammation which should trigger antiviral immunity leading to clearance. Two major families of transcription factors responsible for driving the immune response to viruses are the NF-κB and the interferon regulatory factor (IRF) families. While NF-κB broadly drives pro-inflammatory gene expression and IRFs chiefly drive interferon induction, both collaborate in transactivating many of the same genes in a concerted immune response to viral infection. Here, we report that the MCV protein MC089 specifically inhibits IRF activation from both DNA- and RNA-sensing pathways, making it the first characterized MCV inhibitor to selectively target IRF activation to date. MC089 interacts with proteins required for IRF activation, namely IKKε, TBKBP1 and NAP1. Additionally, MC089 targets RNA sensing by associating with the RNA-sensing adaptor protein mitochondrial antiviral-signalling protein on mitochondria. MC089 displays specificity in its inhibition of IRF3 activation by suppressing immunostimulatory nucleic acid-induced serine 396 phosphorylation without affecting the phosphorylation of serine 386. The selective interaction of MC089 with IRF-regulatory proteins and site-specific inhibition of IRF3 phosphorylation may offer a tool to provide novel insights into the biology of IRF3 regulation.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Yiqi Huang, Valter Bergant, Vincent Grass, Quirin Emslander, M Sabri Hamad, Philipp Hubel, Julia Mergner, Antonio Piras, Karsten Krey, Alexander Henrici, Rupert Öllinger, Yonas M Tesfamariam, Ilaria Dalla Rosa, Till Bunse, Gerd Sutter, Gregor Ebert, Florian I Schmidt, Michael Way, Roland Rad, Andrew G Bowie, Ulrike Protzer, Andreas Pichlmair
Multi-omics characterization of the monkeypox virus infection Journal Article
In: Nat Commun, vol. 15, no. 1, pp. 6778, 2024, ISSN: 2041-1723.
@article{pmid39117661,
title = {Multi-omics characterization of the monkeypox virus infection},
author = {Yiqi Huang and Valter Bergant and Vincent Grass and Quirin Emslander and M Sabri Hamad and Philipp Hubel and Julia Mergner and Antonio Piras and Karsten Krey and Alexander Henrici and Rupert Öllinger and Yonas M Tesfamariam and Ilaria Dalla Rosa and Till Bunse and Gerd Sutter and Gregor Ebert and Florian I Schmidt and Michael Way and Roland Rad and Andrew G Bowie and Ulrike Protzer and Andreas Pichlmair},
doi = {10.1038/s41467-024-51074-6},
issn = {2041-1723},
year = {2024},
date = {2024-08-01},
journal = {Nat Commun},
volume = {15},
number = {1},
pages = {6778},
abstract = {Multiple omics analyzes of Vaccinia virus (VACV) infection have defined molecular characteristics of poxvirus biology. However, little is known about the monkeypox (mpox) virus (MPXV) in humans, which has a different disease manifestation despite its high sequence similarity to VACV. Here, we perform an in-depth multi-omics analysis of the transcriptome, proteome, and phosphoproteome signatures of MPXV-infected primary human fibroblasts to gain insights into the virus-host interplay. In addition to expected perturbations of immune-related pathways, we uncover regulation of the HIPPO and TGF-β pathways. We identify dynamic phosphorylation of both host and viral proteins, which suggests that MAPKs are key regulators of differential phosphorylation in MPXV-infected cells. Among the viral proteins, we find dynamic phosphorylation of H5 that influenced the binding of H5 to dsDNA. Our extensive dataset highlights signaling events and hotspots perturbed by MPXV, extending the current knowledge on poxviruses. We use integrated pathway analysis and drug-target prediction approaches to identify potential drug targets that affect virus growth. Functionally, we exemplify the utility of this approach by identifying inhibitors of MTOR, CHUK/IKBKB, and splicing factor kinases with potent antiviral efficacy against MPXV and VACV.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Christoph Gruber, Lea Krautner, Valter Bergant, Vincent Grass, Zhe Ma, Lara Rheinemann, Ariane Krus, Friederike Reinhardt, Lyupka Mazneykova, Marianne Rocha-Hasler, Dong-Jiunn Jeffery Truong, Gil Gregor Westmeyer, Andreas Pichlmair, Gregor Ebert, Florian Giesert, Wolfgang Wurst
Engineered, nucleocytoplasmic shuttling Cas13d enables highly efficient cytosolic RNA targeting Journal Article
In: Cell Discov, vol. 10, no. 1, pp. 42, 2024, ISSN: 2056-5968.
@article{pmid38609360,
title = {Engineered, nucleocytoplasmic shuttling Cas13d enables highly efficient cytosolic RNA targeting},
author = {Christoph Gruber and Lea Krautner and Valter Bergant and Vincent Grass and Zhe Ma and Lara Rheinemann and Ariane Krus and Friederike Reinhardt and Lyupka Mazneykova and Marianne Rocha-Hasler and Dong-Jiunn Jeffery Truong and Gil Gregor Westmeyer and Andreas Pichlmair and Gregor Ebert and Florian Giesert and Wolfgang Wurst},
doi = {10.1038/s41421-024-00672-1},
issn = {2056-5968},
year = {2024},
date = {2024-04-01},
urldate = {2024-04-01},
journal = {Cell Discov},
volume = {10},
number = {1},
pages = {42},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Constanze A. Jakwerth, Vincent Grass, Anna Erb, Andreas Pichlmair, Georg Boonen, Veronika Butterweck, Carsten B. Schmidt-Weber
Inhibition of SARS-CoV-2 infection and replication by Petasites hybridus CO2-extract (Ze 339) Journal Article
In: Biomedicine & Pharmacotherapy, vol. 170, 2024, ISSN: 0753-3322.
@article{Jakwerth2024,
title = {Inhibition of SARS-CoV-2 infection and replication by Petasites hybridus CO2-extract (Ze 339)},
author = {Constanze A. Jakwerth and Vincent Grass and Anna Erb and Andreas Pichlmair and Georg Boonen and Veronika Butterweck and Carsten B. Schmidt-Weber},
doi = {10.1016/j.biopha.2023.115959},
issn = {0753-3322},
year = {2024},
date = {2024-01-00},
journal = {Biomedicine & Pharmacotherapy},
volume = {170},
publisher = {Elsevier BV},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Solène Denolly, Alexey Stukalov, Uladzimir Barayeu, Alina N. Rosinski, Paraskevi Kritsiligkou, Sebastian Joecks, Tobias P. Dick, Andreas Pichlmair, Ralf Bartenschlager
Zika virus remodelled ER membranes contain proviral factors involved in redox and methylation pathways Journal Article
In: Nat Commun, vol. 14, no. 1, 2023, ISSN: 2041-1723.
@article{Denolly2023b,
title = {Zika virus remodelled ER membranes contain proviral factors involved in redox and methylation pathways},
author = {Solène Denolly and Alexey Stukalov and Uladzimir Barayeu and Alina N. Rosinski and Paraskevi Kritsiligkou and Sebastian Joecks and Tobias P. Dick and Andreas Pichlmair and Ralf Bartenschlager},
doi = {10.1038/s41467-023-43665-6},
issn = {2041-1723},
year = {2023},
date = {2023-12-00},
journal = {Nat Commun},
volume = {14},
number = {1},
publisher = {Springer Science and Business Media LLC},
abstract = {Abstract Zika virus (ZIKV) has emerged as a global health issue, yet neither antiviral therapy nor a vaccine are available. ZIKV is an enveloped RNA virus, replicating in the cytoplasm in close association with ER membranes. Here, we isolate ER membranes from ZIKV-infected cells and determine their proteome. Forty-six host cell factors are enriched in ZIKV remodeled membranes, several of these having a role in redox and methylation pathways. Four proteins are characterized in detail: thioredoxin reductase 1 (TXNRD1) contributing to folding of disulfide bond containing proteins and modulating ZIKV secretion; aldo-keto reductase family 1 member C3 (AKR1C3), regulating capsid protein abundance and thus, ZIKV assembly; biliverdin reductase B (BLVRB) involved in ZIKV induced lipid peroxidation and increasing stability of viral transmembrane proteins; adenosylhomocysteinase (AHCY) indirectly promoting m6 A methylation of ZIKV RNA by decreasing the level of S- adenosyl homocysteine and thus, immune evasion. These results highlight the involvement of redox and methylation enzymes in the ZIKV life cycle and their accumulation at virally remodeled ER membranes. },
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Line Lykke Andersen, Yiqi Huang, Christian Urban, Lila Oubraham, Elena Winheim, Che Stafford, Dennis Nagl, Fionan O'Duill, Thomas Ebert, Thomas Engleitner, Søren Riis Paludan, Anne Krug, Roland Rad, Veit Hornung, Andreas Pichlmair
Systematic
In: The EMBO Journal, vol. 42, no. 23, 2023, ISSN: 1460-2075.
@article{Andersen2023,
title = {Systematic P2Y receptor survey identifies P2Y11 as modulator of immune responses and virus replication in macrophages},
author = {Line Lykke Andersen and Yiqi Huang and Christian Urban and Lila Oubraham and Elena Winheim and Che Stafford and Dennis Nagl and Fionan O'Duill and Thomas Ebert and Thomas Engleitner and Søren Riis Paludan and Anne Krug and Roland Rad and Veit Hornung and Andreas Pichlmair},
doi = {10.15252/embj.2022113279},
issn = {1460-2075},
year = {2023},
date = {2023-12-00},
journal = {The EMBO Journal},
volume = {42},
number = {23},
publisher = {Springer Science and Business Media LLC},
abstract = {Abstract The immune system is in place to assist in ensuring tissue homeostasis, which can be easily perturbed by invading pathogens or nonpathogenic stressors causing tissue damage. Extracellular nucleotides are well known to contribute to innate immune signaling specificity and strength, but how their signaling is relayed downstream of cell surface receptors and how this translates into antiviral immunity is only partially understood. Here, we systematically investigated the responses of human macrophages to extracellular nucleotides, focusing on the nucleotide‐sensing GPRC receptors of the P2Y family. Time‐resolved transcriptomic analysis showed that adenine‐ and uridine‐based nucleotides induce a specific, immediate, and transient cytokine response through the MAPK signaling pathway that regulates transcriptional activation by AP‐1. Using receptor trans‐complementation, we identified a subset of P2Ys (P2Y1, P2Y2, P2Y6, and P2Y11) that govern inflammatory responses via cytokine induction, while others (P2Y4, P2Y11, P2Y12, P2Y13, and P2Y14) directly induce antiviral responses. Notably, P2Y11 combined both activities, and depletion or inhibition of this receptor in macrophages impaired both inflammatory and antiviral responses. Collectively, these results highlight the underappreciated functions of P2Y receptors in innate immune processes. },
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Valter Bergant, Daniel Schnepf, Niklas de Andrade Krätzig, Philipp Hubel, Christian Urban, Thomas Engleitner, Ronald Dijkman, Bernhard Ryffel, Katja Steiger, Percy A. Knolle, Georg Kochs, Roland Rad, Peter Staeheli, Andreas Pichlmair
mRNA 3’UTR lengthening by alternative polyadenylation attenuates inflammatory responses and correlates with virulence of Influenza A virus Journal Article
In: Nat Commun, vol. 14, no. 1, 2023, ISSN: 2041-1723.
@article{Bergant2023,
title = {mRNA 3’UTR lengthening by alternative polyadenylation attenuates inflammatory responses and correlates with virulence of Influenza A virus},
author = {Valter Bergant and Daniel Schnepf and Niklas de Andrade Krätzig and Philipp Hubel and Christian Urban and Thomas Engleitner and Ronald Dijkman and Bernhard Ryffel and Katja Steiger and Percy A. Knolle and Georg Kochs and Roland Rad and Peter Staeheli and Andreas Pichlmair},
doi = {10.1038/s41467-023-40469-6},
issn = {2041-1723},
year = {2023},
date = {2023-12-00},
journal = {Nat Commun},
volume = {14},
number = {1},
publisher = {Springer Science and Business Media LLC},
abstract = {Abstract Changes of mRNA 3’UTRs by alternative polyadenylation (APA) have been associated to numerous pathologies, but the mechanisms and consequences often remain enigmatic. By combining transcriptomics, proteomics and recombinant viruses we show that all tested strains of IAV, including A/PR/8/34(H1N1) (PR8) and A/Cal/07/2009 (H1N1) (Cal09), cause APA. We mapped the effect to the highly conserved glycine residue at position 184 (G184) of the viral non-structural protein 1 (NS1). Unbiased mass spectrometry-based analyses indicate that NS1 causes APA by perturbing the function of CPSF4 and that this function is unrelated to virus-induced transcriptional shutoff. Accordingly, IAV strain PR8, expressing an NS1 variant with weak CPSF binding, does not induce host shutoff but only APA. However, recombinant IAV (PR8) expressing NS1(G184R) lacks binding to CPSF4 and thereby also the ability to cause APA. Functionally, the impaired ability to induce APA leads to an increased inflammatory cytokine production and an attenuated phenotype in a mouse infection model. Investigating diverse viral infection models showed that APA induction is a frequent ability of many pathogens. Collectively, we propose that targeting of the CPSF complex, leading to widespread alternative polyadenylation of host transcripts, constitutes a general immunevasion mechanism employed by a variety of pathogenic viruses. },
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Solène Denolly, Hongbo Guo, Miriam Martens, Anna Płaszczyca, Pietro Scaturro, Vibhu Prasad, Kessiri Kongmanas, Nuntaya Punyadee, Adisak Songjaeng, Dumrong Mairiang, Andreas Pichlmair, Panisadee Avirutnan, Ralf Bartenschlager
In: mBio, vol. 14, no. 5, 2023, ISSN: 2150-7511.
@article{Denolly2023,
title = {Dengue virus NS1 secretion is regulated via importin-subunit β1 controlling expression of the chaperone GRp78 and targeted by the clinical drug ivermectin},
author = {Solène Denolly and Hongbo Guo and Miriam Martens and Anna Płaszczyca and Pietro Scaturro and Vibhu Prasad and Kessiri Kongmanas and Nuntaya Punyadee and Adisak Songjaeng and Dumrong Mairiang and Andreas Pichlmair and Panisadee Avirutnan and Ralf Bartenschlager},
editor = {Sara Cherry and John Schoggins},
doi = {10.1128/mbio.01441-23},
issn = {2150-7511},
year = {2023},
date = {2023-10-31},
journal = {mBio},
volume = {14},
number = {5},
publisher = {American Society for Microbiology},
abstract = {ABSTRACT
Dengue virus (DENV) is a major human pathogen. An important pathogenicity factor is non-structural protein 1 (NS1) required for viral replication and secreted from infected cells. A clinical study indicated that the anti-parasitic drug ivermectin lowers NS1 blood levels without affecting viremia. Ivermectin targets nuclear transport by binding to importin-α, but how NS1 secretion in patients is suppressed by this drug is unknown. We show that ivermectin impairs folding and secretion of endoplasmic reticulum-luminal glycoproteins, including NS1. Proteomic analysis identified chaperones interacting with NS1, including GRp78 (78-kDa glucose-regulated protein, also known as HSPA5 or BIP). This chaperone increased in abundance on DENV infection via activation of the unfolded protein response (UPR). Ivermectin blocked the nuclear transport of transcription factors required for UPR, thus impairing GRp78 upregulation and NS1 secretion. Reduction of GRp78 and NS1 secretion was also observed in patients treated with ivermectin. These results link nuclear transport and its inhibition by ivermectin to folding and secretion of luminal glycoproteins, including DENV NS1.
IMPORTANCE
Dengue virus (DENV) is a major human pathogen that can cause hemorrhagic fever and shock syndrome. One important factor of DENV pathogenicity is non-structural protein 1 (NS1), a glycoprotein that is secreted from infected cells. Here we study the mode of action of the widely used drug ivermectin, used to treat parasitic infections and recently shown to lower NS1 blood levels in DENV-infected patients. We found that ivermectin blocks the nuclear transport of transcription factors required for the expression of chaperones that support the folding and secretion of glycoproteins, including NS1. Impairing nuclear transport of these transcription factors by ivermectin or depleting them from infected cells dampens NS1 folding and thus its secretion. These results reveal a novel mode of action of ivermectin that might apply to other flaviviruses as well.
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Nora Schmidt, Sabina Ganskih, Yuanjie Wei, Alexander Gabel, Sebastian Zielinski, Hasmik Keshishian, Caleb A. Lareau, Liv Zimmermann, Jana Makroczyova, Cadence Pearce, Karsten Krey, Thomas Hennig, Sebastian Stegmaier, Lambert Moyon, Marc Horlacher, Simone Werner, Jens Aydin, Marco Olguin-Nava, Ramya Potabattula, Anuja Kibe, Lars Dölken, Redmond P. Smyth, Neva Caliskan, Annalisa Marsico, Christine Krempl, Jochen Bodem, Andreas Pichlmair, Steven A. Carr, Petr Chlanda, Florian Erhard, Mathias Munschauer
SND1 binds SARS-CoV-2 negative-sense RNA and promotes viral RNA synthesis through NSP9 Journal Article
In: Cell, vol. 186, no. 22, pp. 4834–4850.e23, 2023, ISSN: 0092-8674.
@article{Schmidt2023,
title = {SND1 binds SARS-CoV-2 negative-sense RNA and promotes viral RNA synthesis through NSP9},
author = {Nora Schmidt and Sabina Ganskih and Yuanjie Wei and Alexander Gabel and Sebastian Zielinski and Hasmik Keshishian and Caleb A. Lareau and Liv Zimmermann and Jana Makroczyova and Cadence Pearce and Karsten Krey and Thomas Hennig and Sebastian Stegmaier and Lambert Moyon and Marc Horlacher and Simone Werner and Jens Aydin and Marco Olguin-Nava and Ramya Potabattula and Anuja Kibe and Lars Dölken and Redmond P. Smyth and Neva Caliskan and Annalisa Marsico and Christine Krempl and Jochen Bodem and Andreas Pichlmair and Steven A. Carr and Petr Chlanda and Florian Erhard and Mathias Munschauer},
doi = {10.1016/j.cell.2023.09.002},
issn = {0092-8674},
year = {2023},
date = {2023-10-00},
journal = {Cell},
volume = {186},
number = {22},
pages = {4834--4850.e23},
publisher = {Elsevier BV},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Sandy S Burkart, Darius Schweinoch, Jamie Frankish, Carola Sparn, Sandra Wüst, Christian Urban, Marta Merlo, Vladimir G Magalhães, Antonio Piras, Andreas Pichlmair, Joschka Willemsen, Lars Kaderali, Marco Binder
High-resolution kinetic characterization of the RIG-I-signaling pathway and the antiviral response Journal Article
In: Life Sci. Alliance, vol. 6, no. 10, 2023, ISSN: 2575-1077.
@article{Burkart2023,
title = {High-resolution kinetic characterization of the RIG-I-signaling pathway and the antiviral response},
author = {Sandy S Burkart and Darius Schweinoch and Jamie Frankish and Carola Sparn and Sandra Wüst and Christian Urban and Marta Merlo and Vladimir G Magalhães and Antonio Piras and Andreas Pichlmair and Joschka Willemsen and Lars Kaderali and Marco Binder},
doi = {10.26508/lsa.202302059},
issn = {2575-1077},
year = {2023},
date = {2023-10-00},
journal = {Life Sci. Alliance},
volume = {6},
number = {10},
publisher = {Life Science Alliance, LLC},
abstract = {RIG-I recognizes viral dsRNA and activates a cell-autonomous antiviral response. Upon stimulation, it triggers a signaling cascade leading to the production of type I and III IFNs. IFNs are secreted and signal to elicit the expression of IFN-stimulated genes, establishing an antiviral state of the cell. The topology of this pathway has been studied intensively, however, its exact dynamics are less understood. Here, we employed electroporation to synchronously activate RIG-I, enabling us to characterize cell-intrinsic innate immune signaling at a high temporal resolution. Employing IFNAR1/IFNLR-deficient cells, we could differentiate primary RIG-I signaling from secondary signaling downstream of the IFN receptors. Based on these data, we developed a comprehensive mathematical model capable of simulating signaling downstream of dsRNA recognition by RIG-I and the feedback and signal amplification by IFN. We further investigated the impact of viral antagonists on signaling dynamics. Our work provides a comprehensive insight into the signaling events that occur early upon virus infection and opens new avenues to study and disentangle the complexity of the host–virus interface. },
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Apurva A. Govande, Aleksandra W. Babnis, Christian Urban, Matthias Habjan, Rune Hartmann, Philip J. Kranzusch, Andreas Pichlmair
RNase L-activating 2′–5′ oligoadenylates bind ABCF1, ABCF3 and Decr-1 Journal Article
In: vol. 104, no. 9, 2023, ISSN: 1465-2099.
@article{Govande2023,
title = {RNase L-activating 2′–5′ oligoadenylates bind ABCF1, ABCF3 and Decr-1},
author = {Apurva A. Govande and Aleksandra W. Babnis and Christian Urban and Matthias Habjan and Rune Hartmann and Philip J. Kranzusch and Andreas Pichlmair},
doi = {10.1099/jgv.0.001890},
issn = {1465-2099},
year = {2023},
date = {2023-09-07},
volume = {104},
number = {9},
publisher = {Microbiology Society},
abstract = {A notable signalling mechanism employed by mammalian innate immune signalling pathways uses nucleotide-based second messengers such as 2′3′-cGAMP and 2′–5′-oligoadenylates (OAs), which bind and activate STING and RNase L, respectively. Interestingly, the involvement of nucleotide second messengers to activate antiviral responses is evolutionarily conserved, as evidenced by the identification of an antiviral cGAMP-dependent pathway in Drosophila . Using a mass spectrometry approach, we identified several members of the ABCF family in human, mouse and Drosophila cell lysates as 2′–5′ OA-binding proteins, suggesting an evolutionarily conserved function. Biochemical characterization of these interactions demonstrates high-affinity binding of 2′–5′ OA to ABCF1, dependent on phosphorylated 2′–5′ OA and an intact Walker A/B motif of the ABC cassette of ABCF1. As further support for species-specific interactions with 2′–5′ OA, we additionally identified that the metabolic enzyme Decr1 from mouse, but not human or Drosophila cells, forms a high-affinity complex with 2′–5′ OA. A 1.4 Å co-crystal structure of the mouse Decr1–2′–5′ OA complex explains high-affinity recognition of 2′–5′ OA and the mechanism of species specificity. Despite clear evidence of physical interactions, we could not identify profound antiviral functions of ABCF1, ABCF3 or Decr1 or 2′–5′ OA-dependent regulation of cellular translation rates, as suggested by the engagement of ABCF proteins. Thus, although the biological consequences of the here identified interactions need to be further studied, our data suggest that 2′–5′ OA can serve as a signalling hub to distribute a signal to different recipient proteins. },
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Quirin Emslander, Karsten Krey, Sabri Hamad, Susanne Maidl, Lila Oubraham, Joshua Hesse, Alexander Henrici, Katharina Austen, Julia Mergner, Vincent Grass, Andreas Pichlmair
MDM2 Influences ACE2 Stability and SARS-CoV-2 Uptake Journal Article
In: Viruses, vol. 15, no. 8, 2023, ISSN: 1999-4915.
@article{Emslander2023,
title = {MDM2 Influences ACE2 Stability and SARS-CoV-2 Uptake},
author = {Quirin Emslander and Karsten Krey and Sabri Hamad and Susanne Maidl and Lila Oubraham and Joshua Hesse and Alexander Henrici and Katharina Austen and Julia Mergner and Vincent Grass and Andreas Pichlmair},
doi = {10.3390/v15081763},
issn = {1999-4915},
year = {2023},
date = {2023-08-00},
journal = {Viruses},
volume = {15},
number = {8},
publisher = {MDPI AG},
abstract = {Angiotensin-converting enzyme 2 (ACE2) is the central entry receptor for SARS-CoV-2. However, surprisingly little is known about the effects of host regulators on ACE2 localization, expression, and the associated influence on SARS-CoV-2 infection. Here we identify that ACE2 expression levels are regulated by the E3 ligase MDM2 and that MDM2 levels indirectly influence infection with SARS-CoV-2. Genetic depletion of MDM2 elevated ACE2 expression levels, which strongly promoted infection with all SARS-CoV-2 isolates tested. SARS-CoV-2 spike-pseudotyped viruses and the uptake of non-replication-competent virus-like particles showed that MDM2 affects the viral uptake process. MDM2 ubiquitinates Lysine 788 of ACE2 to induce proteasomal degradation, and degradation of this residue led to higher ACE2 expression levels and superior virus particle uptake. Our study illustrates that cellular regulators of ACE2 stability, such as MDM2, play an important role in defining the infection capabilities of SARS-CoV-2. },
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Thomas Phelan, Clara Lawler, Andreas Pichlmair, Mark A. Little, Andrew G. Bowie, Gareth Brady
Molluscum Contagiosum Virus Protein MC008 Targets NF-κB Activation by Inhibiting Ubiquitination of NEMO Journal Article
In: J Virol, vol. 97, no. 3, 2023, ISSN: 1098-5514.
@article{Phelan2023,
title = {Molluscum Contagiosum Virus Protein MC008 Targets NF-κB Activation by Inhibiting Ubiquitination of NEMO},
author = {Thomas Phelan and Clara Lawler and Andreas Pichlmair and Mark A. Little and Andrew G. Bowie and Gareth Brady},
editor = {Derek Walsh},
doi = {10.1128/jvi.00108-23},
issn = {1098-5514},
year = {2023},
date = {2023-03-30},
journal = {J Virol},
volume = {97},
number = {3},
publisher = {American Society for Microbiology},
abstract = {Inflammation lies at the heart of most human diseases. Understanding the pathways that drive this response is the key to new anti-inflammatory therapies. },
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Yuta Tsukamoto, Takahiro Hiono, Shintaro Yamada, Keita Matsuno, Aileen Faist, Tobias Claff, Jianyu Hou, Vigneshwaran Namasivayam, Anja vom Hemdt, Satoko Sugimoto, Jin Ying Ng, Maria H. Christensen, Yonas M. Tesfamariam, Steven Wolter, Stefan Juranek, Thomas Zillinger, Stefan Bauer, Takatsugu Hirokawa, Florian I. Schmidt, Georg Kochs, Masayuki Shimojima, Yi-Shuian Huang, Andreas Pichlmair, Beate M. Kümmerer, Yoshihiro Sakoda, Martin Schlee, Linda Brunotte, Christa E. Müller, Manabu Igarashi, Hiroki Kato
Inhibition of cellular RNA methyltransferase abrogates influenza virus capping and replication Journal Article
In: Science, vol. 379, no. 6632, pp. 586–591, 2023, ISSN: 1095-9203.
@article{Tsukamoto2023,
title = {Inhibition of cellular RNA methyltransferase abrogates influenza virus capping and replication},
author = {Yuta Tsukamoto and Takahiro Hiono and Shintaro Yamada and Keita Matsuno and Aileen Faist and Tobias Claff and Jianyu Hou and Vigneshwaran Namasivayam and Anja vom Hemdt and Satoko Sugimoto and Jin Ying Ng and Maria H. Christensen and Yonas M. Tesfamariam and Steven Wolter and Stefan Juranek and Thomas Zillinger and Stefan Bauer and Takatsugu Hirokawa and Florian I. Schmidt and Georg Kochs and Masayuki Shimojima and Yi-Shuian Huang and Andreas Pichlmair and Beate M. Kümmerer and Yoshihiro Sakoda and Martin Schlee and Linda Brunotte and Christa E. Müller and Manabu Igarashi and Hiroki Kato},
doi = {10.1126/science.add0875},
issn = {1095-9203},
year = {2023},
date = {2023-02-10},
journal = {Science},
volume = {379},
number = {6632},
pages = {586--591},
publisher = {American Association for the Advancement of Science (AAAS)},
abstract = {
Orthomyxo- and bunyaviruses steal the 5′ cap portion of host RNAs to prime their own transcription in a process called “cap snatching.” We report that RNA modification of the cap portion by host 2′-O-ribose methyltransferase 1 (MTr1) is essential for the initiation of influenza A and B virus replication, but not for other cap-snatching viruses. We identified with in silico compound screening and functional analysis a derivative of a natural product from
Streptomyces
, called trifluoromethyl-tubercidin (TFMT), that inhibits MTr1 through interaction at its
S
-adenosyl-
l
-methionine binding pocket to restrict influenza virus replication. Mechanistically, TFMT impairs the association of host cap RNAs with the viral polymerase basic protein 2 subunit in human lung explants and in vivo in mice. TFMT acts synergistically with approved anti-influenza drugs.
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Knud Esser, Xiaoming Cheng, Jochen M. Wettengel, Julie Lucifora, Lea Hansen-Palmus, Katharina Austen, Armando A. Roca Suarez, Sarah Heintz, Barbara Testoni, Firat Nebioglu, Minh Tu Pham, Shangqing Yang, Alma Zernecke, Dirk Wohlleber, Marc Ringelhan, Mathias Broxtermann, Daniel Hartmann, Norbert Hüser, Julia Mergner, Andreas Pichlmair, Wolfgang E. Thasler, Mathias Heikenwalder, Georg Gasteiger, Andreas Blutke, Axel Walch, Percy A. Knolle, Ralf Bartenschlager, Ulrike Protzer
Hepatitis B Virus Targets Lipid Transport Pathways to Infect Hepatocytes Journal Article
In: Cellular and Molecular Gastroenterology and Hepatology, vol. 16, no. 2, pp. 201–221, 2023, ISSN: 2352-345X.
@article{Esser2023,
title = {Hepatitis B Virus Targets Lipid Transport Pathways to Infect Hepatocytes},
author = {Knud Esser and Xiaoming Cheng and Jochen M. Wettengel and Julie Lucifora and Lea Hansen-Palmus and Katharina Austen and Armando A. Roca Suarez and Sarah Heintz and Barbara Testoni and Firat Nebioglu and Minh Tu Pham and Shangqing Yang and Alma Zernecke and Dirk Wohlleber and Marc Ringelhan and Mathias Broxtermann and Daniel Hartmann and Norbert Hüser and Julia Mergner and Andreas Pichlmair and Wolfgang E. Thasler and Mathias Heikenwalder and Georg Gasteiger and Andreas Blutke and Axel Walch and Percy A. Knolle and Ralf Bartenschlager and Ulrike Protzer},
doi = {10.1016/j.jcmgh.2023.03.011},
issn = {2352-345X},
year = {2023},
date = {2023-00-00},
journal = {Cellular and Molecular Gastroenterology and Hepatology},
volume = {16},
number = {2},
pages = {201--221},
publisher = {Elsevier BV},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Laura M. Mateyka, Vincent Grass, Andreas Pichlmair, Dirk H. Busch, Elvira D’Ippolito
SARS-CoV-2 CD8+ T cell killing assays using replicating viruses and transgenic antigens Journal Article
In: STAR Protocols, vol. 3, no. 4, 2022, ISSN: 2666-1667.
@article{Mateyka2022,
title = {SARS-CoV-2 CD8+ T cell killing assays using replicating viruses and transgenic antigens},
author = {Laura M. Mateyka and Vincent Grass and Andreas Pichlmair and Dirk H. Busch and Elvira D’Ippolito},
doi = {10.1016/j.xpro.2022.101699},
issn = {2666-1667},
year = {2022},
date = {2022-12-00},
journal = {STAR Protocols},
volume = {3},
number = {4},
publisher = {Elsevier BV},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Valter Bergant, Shintaro Yamada, Vincent Grass, Yuta Tsukamoto, Teresa Lavacca, Karsten Krey, Maria‐Teresa Mühlhofer, Sabine Wittmann, Armin Ensser, Alexandra Herrmann, Anja vom Hemdt, Yuriko Tomita, Shutoku Matsuyama, Takatsugu Hirokawa, Yiqi Huang, Antonio Piras, Constanze A Jakwerth, Madlen Oelsner, Susanne Thieme, Alexander Graf, Stefan Krebs, Helmut Blum, Beate M Kümmerer, Alexey Stukalov, Carsten B Schmidt‐Weber, Manabu Igarashi, Thomas Gramberg, Andreas Pichlmair, Hiroki Kato
Attenuation of
In: The EMBO Journal, vol. 41, no. 17, 2022, ISSN: 1460-2075.
@article{Bergant2022,
title = {Attenuation of SARS‐CoV ‐2 replication and associated inflammation by concomitant targeting of viral and host cap 2'‐O‐ribose methyltransferases},
author = {Valter Bergant and Shintaro Yamada and Vincent Grass and Yuta Tsukamoto and Teresa Lavacca and Karsten Krey and Maria‐Teresa Mühlhofer and Sabine Wittmann and Armin Ensser and Alexandra Herrmann and Anja vom Hemdt and Yuriko Tomita and Shutoku Matsuyama and Takatsugu Hirokawa and Yiqi Huang and Antonio Piras and Constanze A Jakwerth and Madlen Oelsner and Susanne Thieme and Alexander Graf and Stefan Krebs and Helmut Blum and Beate M Kümmerer and Alexey Stukalov and Carsten B Schmidt‐Weber and Manabu Igarashi and Thomas Gramberg and Andreas Pichlmair and Hiroki Kato},
doi = {10.15252/embj.2022111608},
issn = {1460-2075},
year = {2022},
date = {2022-09-00},
journal = {The EMBO Journal},
volume = {41},
number = {17},
publisher = {Springer Science and Business Media LLC},
abstract = {Abstract The SARS‐CoV‐2 infection cycle is a multistage process that relies on functional interactions between the host and the pathogen. Here, we repurposed antiviral drugs against both viral and host enzymes to pharmaceutically block methylation of the viral RNA 2'‐O‐ribose cap needed for viral immune escape. We find that the host cap 2'‐O‐ribose methyltransferase MTr1 can compensate for loss of viral NSP16 methyltransferase in facilitating virus replication. Concomitant inhibition of MTr1 and NSP16 efficiently suppresses SARS‐CoV‐2 replication. Using in silico target‐based drug screening, we identify a bispecific MTr1/NSP16 inhibitor with anti‐SARS‐CoV‐2 activity in vitro and in vivo but with unfavorable side effects. We further show antiviral activity of inhibitors that target independent stages of the host SAM cycle providing the methyltransferase co‐substrate. In particular, the adenosylhomocysteinase (AHCY) inhibitor DZNep is antiviral in in vitro , in ex vivo , and in a mouse infection model and synergizes with existing COVID‐19 treatments. Moreover, DZNep exhibits a strong immunomodulatory effect curbing infection‐induced hyperinflammation and reduces lung fibrosis markers ex vivo . Thus, multispecific and metabolic MTase inhibitors constitute yet unexplored treatment options against COVID‐19. },
keywords = {},
pubstate = {published},
tppubtype = {article}
}
M.I. Hollenhorst, R. Nandigama, S.B. Evers, I. Gamayun, N.A. Wadood, A. Salah, M. Pieper, A. Wyatt, A. Stukalov, A. Gebhardt, W. Nadolni, W. Burow, C. Herr, C. Beisswenger, S. Kusumakshi, F. Ectors, T.I. Kichko, L. Hübner, P. Reeh, A. Munder, S. Wienhold, M. Witzenrath, R. Bals, V. Flockerzi, T. Gudermann, M. Bischoff , P. Lipp, S. Zierler, V. Chubanov, A. Pichlmair, P. König, U. Boehm, G. Krasteva-Christ
Bitter Taste Signaling in Tracheal Epithelial Brush Cells Elicits Innate Immune Responses to Bacterial Infection Journal Article
In: The Journal of Clinical Investigation, vol. 132, no. 13, 2022, ISSN: 1558-8238.
@article{Krasteva-Christ2022,
title = {Bitter Taste Signaling in Tracheal Epithelial Brush Cells Elicits Innate Immune Responses to Bacterial Infection},
author = {M.I. Hollenhorst, R. Nandigama, S.B. Evers, I. Gamayun, N.A. Wadood, A. Salah, M. Pieper, A. Wyatt, A. Stukalov, A. Gebhardt, W. Nadolni, W. Burow, C. Herr, C. Beisswenger, S. Kusumakshi, F. Ectors, T.I. Kichko,
L. Hübner, P. Reeh, A. Munder, S. Wienhold, M. Witzenrath, R. Bals, V. Flockerzi, T. Gudermann, M. Bischoff , P. Lipp, S. Zierler, V. Chubanov, A. Pichlmair, P. König, U. Boehm, G. Krasteva-Christ},
url = {https://doi.org/10.1172/JCI150951},
doi = {10.1172/JCI150951},
issn = {1558-8238},
year = {2022},
date = {2022-07-01},
journal = {The Journal of Clinical Investigation},
volume = {132},
number = {13},
abstract = {Constant exposure of the airways to inhaled pathogens requires efficient early immune responses protecting against infections. How bacteria on the epithelial surface are detected and first-line protective mechanisms are initiated are not well understood. We have recently shown that tracheal brush cells (BCs) express functional taste receptors. Here we report that bitter taste signaling in murine BCs induces neurogenic inflammation. We demonstrate that BC signaling stimulates adjacent sensory nerve endings in the trachea to release the neuropeptides CGRP and substance P that mediate plasma extravasation, neutrophil recruitment, and diapedesis. Moreover, we show that bitter tasting quorum-sensing molecules from Pseudomonas aeruginosa activate tracheal BCs. BC signaling depends on the key taste transduction gene Trpm5, triggers secretion of immune mediators, among them the most abundant member of the complement system, and is needed to combat P. aeruginosa infections. Our data provide functional insight into first-line defense mechanisms against bacterial infections of the lung.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Hengyuan Liu, Valter Bergant, Goar Frishman, Andreas Ruepp, Andreas Pichlmair, Michelle Vincendeau, Dmitrij Frishman
Influenza A Virus Infection Reactivates Human Endogenous Retroviruses Associated with Modulation of Antiviral Immunity Journal Article
In: Viruses, vol. 14, no. 7, 2022, ISSN: 1999-4915.
@article{Liu2022,
title = {Influenza A Virus Infection Reactivates Human Endogenous Retroviruses Associated with Modulation of Antiviral Immunity},
author = {Hengyuan Liu and Valter Bergant and Goar Frishman and Andreas Ruepp and Andreas Pichlmair and Michelle Vincendeau and Dmitrij Frishman},
doi = {10.3390/v14071591},
issn = {1999-4915},
year = {2022},
date = {2022-07-00},
journal = {Viruses},
volume = {14},
number = {7},
publisher = {MDPI AG},
abstract = {Human endogenous retrovirus (HERVs), normally silenced by methylation or mutations, can be reactivated by multiple environmental factors, including infections with exogenous viruses. In this work, we investigated the transcriptional activity of HERVs in human A549 cells infected by two wild-type (PR8M, SC35M) and one mutated (SC35MΔNS1) strains of Influenza A virus (IAVs). We found that the majority of differentially expressed HERVs (DEHERVS) and genes (DEGs) were up-regulated in the infected cells, with the most significantly enriched biological processes associated with the genes differentially expressed exclusively in SC35MΔNS1 being linked to the immune system. Most DEHERVs in PR8M and SC35M are mammalian apparent LTR retrotransposons, while in SC35MΔNS1, more HERV loci from the HERVW9 group were differentially expressed. Furthermore, up-regulated pairs of HERVs and genes in close chromosomal proximity to each other tended to be associated with immune responses, which implies that specific HERV groups might have the potential to trigger specific gene networks and influence host immunological pathways. },
keywords = {},
pubstate = {published},
tppubtype = {article}
}
M.C. Serrero, V. Girault, S. Weigang, T.M. Greco, A. Ramos-Nascimento, F. Anderson, A. Piras, A.H. Martinez, J. Hertzog, A. Binz, A. Pohlmann, U. Prank, J. Rehwinkel, R. Bauerfeind, I.M. Cristea, A. Pichlmair, G. Kochs, B. Sodeik
The Interferon-inducible GTPase MxB Promotes Capsid Disassembly and Genome Release of Herpesviruses Journal Article
In: eLife, vol. 11, pp. e76804, 2022, ISSN: 2050-084X.
@article{10.7554/eLife.76804,
title = {The Interferon-inducible GTPase MxB Promotes Capsid Disassembly and Genome Release of Herpesviruses},
author = {M.C. Serrero and V. Girault and S. Weigang and T.M. Greco and A. Ramos-Nascimento and F. Anderson and A. Piras and A.H. Martinez and J. Hertzog and A. Binz and A. Pohlmann and U. Prank and J. Rehwinkel and R. Bauerfeind and I.M. Cristea and A. Pichlmair and G. Kochs and B. Sodeik},
editor = {Adam P Geballe and Päivi M Ojala},
url = {https://doi.org/10.7554/eLife.76804},
doi = {10.7554/eLife.76804},
issn = {2050-084X},
year = {2022},
date = {2022-04-01},
journal = {eLife},
volume = {11},
pages = {e76804},
publisher = {eLife Sciences Publications, Ltd},
abstract = {Host proteins sense viral products and induce defence mechanisms, particularly in immune cells. Using cell-free assays and quantitative mass spectrometry, we determined the interactome of capsid-host protein complexes of herpes simplex virus and identified the large dynamin-like GTPase myxovirus resistance protein B (MxB) as an interferon-inducible protein interacting with capsids. Electron microscopy analyses showed that cytosols containing MxB had the remarkable capability to disassemble the icosahedral capsids of herpes simplex viruses and varicella zoster virus into flat sheets of connected triangular faces. In contrast, capsids remained intact in cytosols with MxB mutants unable to hydrolyse GTP or to dimerize. Our data suggest that MxB senses herpesviral capsids, mediates their disassembly, and thereby restricts the efficiency of nuclear targeting of incoming capsids and/or the assembly of progeny capsids. The resulting premature release of viral genomes from capsids may enhance the activation of DNA sensors, and thereby amplify the innate immune responses.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
S. Ambike, C. Cheng, M. Feuerherd, S. Velkov, D. Baldassi, S.Q. Afridi, D. Porras-Gonzalez, X. Wei, P. Hagen, N. Kneidinger, M.G. Stoleriu, V. Grass, G. Burgstaller, A. Pichlmair, O.M. Merkel, C. Ko, T. Michler
Targeting Genomic SARS-CoV-2 RNA with siRNAs Allows Efficient Inhibition of Viral Replication and Spread Journal Article
In: Nucleic Acids Research, vol. 50, no. 1, pp. 333-349, 2022, ISSN: 0305-1048.
@article{10.1093/nar/gkab1248,
title = {Targeting Genomic SARS-CoV-2 RNA with siRNAs Allows Efficient Inhibition of Viral Replication and Spread},
author = {S. Ambike and C. Cheng and M. Feuerherd and S. Velkov and D. Baldassi and S.Q. Afridi and D. Porras-Gonzalez and X. Wei and P. Hagen and N. Kneidinger and M.G. Stoleriu and V. Grass and G. Burgstaller and A. Pichlmair and O.M. Merkel and C. Ko and T. Michler},
url = {https://doi.org/10.1093/nar/gkab1248},
doi = {10.1093/nar/gkab1248},
issn = {0305-1048},
year = {2022},
date = {2022-01-11},
journal = {Nucleic Acids Research},
volume = {50},
number = {1},
pages = {333-349},
abstract = {A promising approach to tackle the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) could be small interfering (si)RNAs. So far it is unclear, which viral replication steps can be efficiently inhibited with siRNAs. Here, we report that siRNAs can target genomic RNA (gRNA) of SARS-CoV-2 after cell entry, and thereby terminate replication before start of transcription and prevent virus-induced cell death. Coronaviruses replicate via negative sense RNA intermediates using a unique discontinuous transcription process. As a result, each viral RNA contains identical sequences at the 5′ and 3′ end. Surprisingly, siRNAs were not active against intermediate negative sense transcripts. Targeting common sequences shared by all viral transcripts allowed simultaneous suppression of gRNA and subgenomic (sg)RNAs by a single siRNA. The most effective suppression of viral replication and spread, however, was achieved by siRNAs that targeted open reading frame 1 (ORF1) which only exists in gRNA. In contrast, siRNAs that targeted the common regions of transcripts were outcompeted by the highly abundant sgRNAs leading to an impaired antiviral efficacy. Verifying the translational relevance of these findings, we show that a chemically modified siRNA that targets a highly conserved region of ORF1, inhibited SARS-CoV-2 replication ex vivo in explants of the human lung. Our work encourages the development of siRNA-based therapies for COVID-19 and suggests that early therapy start, or prophylactic application, together with specifically targeting gRNA, might be key for high antiviral efficacy.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Q. Emslander, K. Vogele, P. Braun, J. Stender, C. Willy, M. Joppich, J.A. Hammerl, M. Abele, C. Meng, A. Pichlmair, C. Ludwig, J.J. Bugert, F.C. Simmel, G.G. Westmeyer
Cell-free Production of Personalized Therapeutic Phages Targeting Multidrug-resistant Bacteria Journal Article
In: Cell Chemical Biology, 2022, ISSN: 2451-9456.
@article{EMSLANDER2022,
title = {Cell-free Production of Personalized Therapeutic Phages Targeting Multidrug-resistant Bacteria},
author = {Q. Emslander and K. Vogele and P. Braun and J. Stender and C. Willy and M. Joppich and J.A. Hammerl and M. Abele and C. Meng and A. Pichlmair and C. Ludwig and J.J. Bugert and F.C. Simmel and G.G. Westmeyer},
url = {https://www.sciencedirect.com/science/article/pii/S2451945622002355},
doi = {https://doi.org/10.1016/j.chembiol.2022.06.003},
issn = {2451-9456},
year = {2022},
date = {2022-01-01},
journal = {Cell Chemical Biology},
abstract = {Bacteriophages are potent therapeutics against biohazardous bacteria, which rapidly develop multidrug resistance. However, routine administration of phage therapy is hampered by a lack of rapid production, safe bioengineering, and detailed characterization of phages. Thus, we demonstrate a comprehensive cell-free platform for personalized production, transient engineering, and proteomic characterization of a broad spectrum of phages. Using mass spectrometry, we validated hypothetical and non-structural proteins and could also monitor the protein expression during phage assembly. Notably, a few microliters of a one-pot reaction produced effective doses of phages against enteroaggregative Escherichia coli (EAEC), Yersinia pestis, and Klebsiella pneumoniae. By co-expressing suitable host factors, we could extend the range of cell-free production to phages targeting gram-positive bacteria. We further introduce a non-genomic phage engineering method, which adds functionalities for only one replication cycle. In summary, we expect this cell-free methodology to foster reverse and forward phage engineering and customized production of clinical-grade bacteriophages.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
G. Jocher, V. Grass, S.K. Tschirner, L. Riepler, S. Breimann, T. Kaya, M. Oelsner, S.M. Hamad, L.I. Hofmann, C.P. Blobel, C.B. Schmidt-Weber, O. Gokce, C.A. Jakwerth, J. Trimpert, J. Kimpel, A. Pichlmair, S.F. Lichtenthaler
ADAM10 and ADAM17 Promote SARS-CoV-2 Cell Entry and Spike Protein-mediated Lung Cell Fusion Journal Article
In: EMBO reports, vol. 23, no. 6, pp. e54305, 2022.
@article{https://doi.org/10.15252/embr.202154305,
title = {ADAM10 and ADAM17 Promote SARS-CoV-2 Cell Entry and Spike Protein-mediated Lung Cell Fusion},
author = {G. Jocher and V. Grass and S.K. Tschirner and L. Riepler and S. Breimann and T. Kaya and M. Oelsner and S.M. Hamad and L.I. Hofmann and C.P. Blobel and C.B. Schmidt-Weber and O. Gokce and C.A. Jakwerth and J. Trimpert and J. Kimpel and A. Pichlmair and S.F. Lichtenthaler},
url = {https://www.embopress.org/doi/abs/10.15252/embr.202154305},
doi = {https://doi.org/10.15252/embr.202154305},
year = {2022},
date = {2022-01-01},
journal = {EMBO reports},
volume = {23},
number = {6},
pages = {e54305},
abstract = {Abstract The severe-acute-respiratory-syndrome-coronavirus-2 (SARS-CoV-2) is the causative agent of COVID-19, but host cell factors contributing to COVID-19 pathogenesis remain only partly understood. We identify the host metalloprotease ADAM17 as a facilitator of SARS-CoV-2 cell entry and the metalloprotease ADAM10 as a host factor required for lung cell syncytia formation, a hallmark of COVID-19 pathology. ADAM10 and ADAM17, which are broadly expressed in the human lung, cleave the SARS-CoV-2 spike protein (S) in vitro, indicating that ADAM10 and ADAM17 contribute to the priming of S, an essential step for viral entry and cell fusion. ADAM protease-targeted inhibitors severely impair lung cell infection by the SARS-CoV-2 variants of concern alpha, beta, delta, and omicron and also reduce SARS-CoV-2 infection of primary human lung cells in a TMPRSS2 protease-independent manner. Our study establishes ADAM10 and ADAM17 as host cell factors for viral entry and syncytia formation and defines both proteases as potential targets for antiviral drug development.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
K.I. Wagner, L.M. Mateyka, S. Jarosch, V. Grass, S. Weber, K. Schober, M. Hammel, T. Burrell, B. Kalali, H. Poppert, H. Beyer, S. Schambeck, S. Holdenrieder, A. Strötges-Achatz, V. Haselmann, M. Neumaier, J. Erber, A. Priller, S. Yazici, H. Roggendorf, M. Odendahl, T. Tonn, A. Dick, K. Witter, H. Mijočević, U. Protzer, P.A. Knolle, A. Pichlmair, C.S. Crowell, M. Gerhard, E. D’Ippolito, D.H. Busch
Recruitment of Highly Cytotoxic CD8+ T Cell Receptors in Mild SARS-CoV-2 Infection Journal Article
In: Cell Reports, vol. 38, no. 2, pp. 110214, 2022, ISSN: 2211-1247.
@article{WAGNER2022110214,
title = {Recruitment of Highly Cytotoxic CD8+ T Cell Receptors in Mild SARS-CoV-2 Infection},
author = {K.I. Wagner and L.M. Mateyka and S. Jarosch and V. Grass and S. Weber and K. Schober and M. Hammel and T. Burrell and B. Kalali and H. Poppert and H. Beyer and S. Schambeck and S. Holdenrieder and A. Strötges-Achatz and V. Haselmann and M. Neumaier and J. Erber and A. Priller and S. Yazici and H. Roggendorf and M. Odendahl and T. Tonn and A. Dick and K. Witter and H. Mijočević and U. Protzer and P.A. Knolle and A. Pichlmair and C.S. Crowell and M. Gerhard and E. D’Ippolito and D.H. Busch},
url = {https://www.sciencedirect.com/science/article/pii/S2211124721017186},
doi = {https://doi.org/10.1016/j.celrep.2021.110214},
issn = {2211-1247},
year = {2022},
date = {2022-01-01},
journal = {Cell Reports},
volume = {38},
number = {2},
pages = {110214},
abstract = {T cell immunity is crucial for control of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and has been studied widely on a quantitative level. However, the quality of responses, in particular of CD8+ T cells, has only been investigated marginally so far. Here, we isolate T cell receptor (TCR) repertoires specific for immunodominant SARS-CoV-2 epitopes restricted to common human Leukocyte antigen (HLA) class I molecules in convalescent individuals. SARS-CoV-2-specific CD8+ T cells are detected up to 12 months after infection. TCR repertoires are diverse, with heterogeneous functional avidity and cytotoxicity toward virus-infected cells, as demonstrated for TCR-engineered T cells. High TCR functionality correlates with gene signatures that, remarkably, could be retrieved for each epitope:HLA combination analyzed. Overall, our data demonstrate that polyclonal and highly functional CD8+ TCRs—classic features of protective immunity—are recruited upon mild SARS-CoV-2 infection, providing tools to assess the quality of and potentially restore functional CD8+ T cell immunity.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
F.L. Pennemann, A. Mussabekova, C. Urban, A. Stukalov, L.L. Andersen, V. Grass, T.M. Lavacca, C. Holze, L. Oubraham, Y. Benamrouche, E. Girardi, R.E. Boulos, R. Hartmann, G. Superti-Furga, M. Habjan, J. Imler, C. Meignin, A. Pichlmair
Cross-species Analysis of Viral Nucleic Acid Interacting Proteins Identifies TAOKs as Innate Immune Regulators Journal Article
In: Nature Communications, vol. 12, no. 1, pp. 7009, 2021, ISSN: 2041-1723.
@article{Pennemann2021,
title = {Cross-species Analysis of Viral Nucleic Acid Interacting Proteins Identifies TAOKs as Innate Immune Regulators},
author = {F.L. Pennemann and A. Mussabekova and C. Urban and A. Stukalov and L.L. Andersen and V. Grass and T.M. Lavacca and C. Holze and L. Oubraham and Y. Benamrouche and E. Girardi and R.E. Boulos and R. Hartmann and G. Superti-Furga and M. Habjan and J. Imler and C. Meignin and A. Pichlmair},
url = {https://doi.org/10.1038/s41467-021-27192-w},
doi = {10.1038/s41467-021-27192-w},
issn = {2041-1723},
year = {2021},
date = {2021-12-01},
journal = {Nature Communications},
volume = {12},
number = {1},
pages = {7009},
abstract = {The cell intrinsic antiviral response of multicellular organisms developed over millions of years and critically relies on the ability to sense and eliminate viral nucleic acids. Here we use an affinity proteomics approach in evolutionary distant species (human, mouse and fly) to identify proteins that are conserved in their ability to associate with diverse viral nucleic acids. This approach shows a core of orthologous proteins targeting viral genetic material and species-specific interactions. Functional characterization of the influence of 181 candidates on replication of 6 distinct viruses in human cells and flies identifies 128 nucleic acid binding proteins with an impact on virus growth. We identify the family of TAO kinases (TAOK1, −2 and −3) as dsRNA-interacting antiviral proteins and show their requirement for type-I interferon induction. Depletion of TAO kinases in mammals or flies leads to an impaired response to virus infection characterized by a reduced induction of interferon stimulated genes in mammals and impaired expression of srg1 and diedel in flies. Overall, our study shows a larger set of proteins able to mediate the interaction between viral genetic material and host factors than anticipated so far, attesting to the ancestral roots of innate immunity and to the lineage-specific pressures exerted by viruses.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
G. Griffante, F. Gugliesi, S. Pasquero, V. DellÓste, M. Biolatti, A.J. Salinger, S. Mondal, P.R. Thompson, E. Weerapana, R.J. Lebbink, J.A. Soppe, T. Stamminger, V. Girault, A. Pichlmair, G. Oroszlán, D.M. Coen, M. De Andrea, S. Landolfo
Human Cytomegalovirus-induced Host Protein Citrullination is Crucial for Viral Replication Journal Article
In: Nature Communications, vol. 12, no. 1, pp. 3910, 2021, ISSN: 20411723.
@article{Griffante2021,
title = {Human Cytomegalovirus-induced Host Protein Citrullination is Crucial for Viral Replication},
author = {G. Griffante and F. Gugliesi and S. Pasquero and V. DellÓste and M. Biolatti and A.J. Salinger and S. Mondal and P.R. Thompson and E. Weerapana and R.J. Lebbink and J.A. Soppe and T. Stamminger and V. Girault and A. Pichlmair and G. Oroszlán and D.M. Coen and M. De Andrea and S. Landolfo},
url = {http://www.nature.com/articles/s41467-021-24178-6},
doi = {10.1038/s41467-021-24178-6},
issn = {20411723},
year = {2021},
date = {2021-12-01},
journal = {Nature Communications},
volume = {12},
number = {1},
pages = {3910},
abstract = {Citrullination is the conversion of arginine-to-citrulline by protein arginine deiminases (PADs), whose dysregulation is implicated in the pathogenesis of various types of cancers and autoimmune diseases. Consistent with the ability of human cytomegalovirus (HCMV) to induce post-translational modifications of cellular proteins to gain a survival advantage, we show that HCMV infection of primary human fibroblasts triggers PAD-mediated citrullination of several host proteins, and that this activity promotes viral fitness. Citrullinome analysis reveals significant changes in deimination levels of both cellular and viral proteins, with interferon (IFN)-inducible protein IFIT1 being among the most heavily deiminated one. As genetic depletion of IFIT1 strongly enhances HCMV growth, and in vitro IFIT1 citrullination impairs its ability to bind to 5'-ppp-RNA, we propose that viral-induced IFIT1 citrullination is a mechanism of HCMV evasion from host antiviral resistance. Overall, our findings point to a crucial role of citrullination in subverting cellular responses to viral infection.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
B.T. Laudenbach, K. Krey, Q. Emslander, L.L. Andersen, A. Reim, P. Scaturro, S. Mundigl, C. Dächert, K. Manske, M. Moser, J. Ludwig, D. Wohlleber, A. Kröger, M. Binder, A. Pichlmair
NUDT2 Initiates Viral RNA Degradation by Removal of 5'-phosphates Journal Article
In: Nature Communications, vol. 12, no. 1, pp. 6918, 2021, ISSN: 2041-1723.
@article{Laudenbach2021,
title = {NUDT2 Initiates Viral RNA Degradation by Removal of 5'-phosphates},
author = {B.T. Laudenbach and K. Krey and Q. Emslander and L.L. Andersen and A. Reim and P. Scaturro and S. Mundigl and C. Dächert and K. Manske and M. Moser and J. Ludwig and D. Wohlleber and A. Kröger and M. Binder and A. Pichlmair},
url = {https://doi.org/10.1038/s41467-021-27239-y},
doi = {10.1038/s41467-021-27239-y},
issn = {2041-1723},
year = {2021},
date = {2021-11-25},
journal = {Nature Communications},
volume = {12},
number = {1},
pages = {6918},
abstract = {While viral replication processes are largely understood, comparably little is known on cellular mechanisms degrading viral RNA. Some viral RNAs bear a 5'-triphosphate (PPP-) group that impairs degradation by the canonical 5textasciiacutex-3textasciiacutex degradation pathway. Here we show that the Nudix hydrolase 2 (NUDT2) trims viral PPP-RNA into monophosphorylated (P)-RNA, which serves as a substrate for the 5'-3' exonuclease XRN1. NUDT2 removes 5'-phosphates from PPP-RNA in an RNA sequence- and overhang-independent manner and its ablation in cells increases growth of PPP-RNA viruses, suggesting an involvement in antiviral immunity. NUDT2 is highly homologous to bacterial RNA pyrophosphatase H (RppH), a protein involved in the metabolism of bacterial mRNA, which is 5'-tri- or diphosphorylated. Our results show a conserved function between bacterial RppH and mammalian NUDT2, indicating that the function may have adapted from a protein responsible for RNA turnover in bacteria into a protein involved in the immune defence in mammals.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
D.S. Fischer, M. Ansari, K.I. Wagner, S. Jarosch, Y. Huang, C.H. Mayr, M. Strunz, N.J. Lang, E. D'Ippolito, M. Hammel, L. Mateyka, S. Weber, L.S. Wolff, K. Witter, I.E. Fernandez, G. Leuschner, K. Milger, M. Frankenberger, L. Nowak, K. Heinig-Menhard, I. Koch, M.G. Stoleriu, A. Hilgendorff, J. Behr, A. Pichlmair, B. Schubert, F.J. Theis, D.H. Busch, H.B. Schiller, K. Schober
Single-cell RNA Sequencing Reveals ex vivo Signatures of SARS-CoV-2-reactive T Cells Through ‘Reverse Phenotyping' Journal Article
In: Nature Communications, vol. 12, no. 1, pp. 4515, 2021, ISSN: 2041-1723.
@article{Fischer2021b,
title = {Single-cell RNA Sequencing Reveals ex vivo Signatures of SARS-CoV-2-reactive T Cells Through ‘Reverse Phenotyping'},
author = {D.S. Fischer and M. Ansari and K.I. Wagner and S. Jarosch and Y. Huang and C.H. Mayr and M. Strunz and N.J. Lang and E. D'Ippolito and M. Hammel and L. Mateyka and S. Weber and L.S. Wolff and K. Witter and I.E. Fernandez and G. Leuschner and K. Milger and M. Frankenberger and L. Nowak and K. Heinig-Menhard and I. Koch and M.G. Stoleriu and A. Hilgendorff and J. Behr and A. Pichlmair and B. Schubert and F.J. Theis and D.H. Busch and H.B. Schiller and K. Schober},
url = {http://www.nature.com/articles/s41467-021-24730-4},
doi = {10.1038/s41467-021-24730-4},
issn = {2041-1723},
year = {2021},
date = {2021-07-26},
journal = {Nature Communications},
volume = {12},
number = {1},
pages = {4515},
abstract = {The in vivo phenotypic profile of T cells reactive to severe acute respiratory syndrome (SARS)-CoV-2 antigens remains poorly understood. Conventional methods to detect antigen-reactive T cells require in vitro antigenic re-stimulation or highly individualized peptide-human leukocyte antigen (pHLA) multimers. Here, we use single-cell RNA sequencing to identify and profile SARS-CoV-2-reactive T cells from Coronavirus Disease 2019 (COVID-19) patients. To do so, we induce transcriptional shifts by antigenic stimulation in vitro and take advantage of natural T cell receptor (TCR) sequences of clonally expanded T cells as barcodes for ‘reverse phenotyping'. This allows identification of SARS-CoV-2-reactive TCRs and reveals phenotypic effects introduced by antigen-specific stimulation. We characterize transcriptional signatures of currently and previously activated SARS-CoV-2-reactive T cells, and show correspondence with phenotypes of T cells from the respiratory tract of patients with severe disease in the presence or absence of virus in independent cohorts. Reverse phenotyping is a powerful tool to provide an integrated insight into cellular states of SARS-CoV-2-reactive T cells across tissues and activation states.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
A. Holleufer, K.G. Winther, H.H. Gad, X. Ai, Y. Chen, L. Li, Z. Wei, H. Deng, J. Liu, N.A. Frederiksen, B. Simonsen, L.L. Andersen, K. Kleigrewe, L. Dalskov, A. Pichlmair, H. Cai, J. Imler, R. Hartmann
Two cGAS-like Receptors Induce Antiviral Immunity in Drosophila Journal Article
In: Nature, 2021, ISSN: 1476-4687.
@article{Holleufer2021,
title = {Two cGAS-like Receptors Induce Antiviral Immunity in Drosophila},
author = {A. Holleufer and K.G. Winther and H.H. Gad and X. Ai and Y. Chen and L. Li and Z. Wei and H. Deng and J. Liu and N.A. Frederiksen and B. Simonsen and L.L. Andersen and K. Kleigrewe and L. Dalskov and A. Pichlmair and H. Cai and J. Imler and R. Hartmann},
url = {https://doi.org/10.1038/s41586-021-03800-z},
doi = {10.1038/s41586-021-03800-z},
issn = {1476-4687},
year = {2021},
date = {2021-07-14},
journal = {Nature},
abstract = {In mammals, cyclic GMP-AMP (cGAMP) synthase (cGAS) produces the cyclic dinucleotide (CDN) 2'3'-cGAMP in response to cytosolic DNA and this triggers an antiviral immune response. cGAS belongs to a large family of cGAS/DncV-like nucleotidyltransferases, present in both prokaryotes1 and eukaryotes2--5. In bacteria, these enzymes synthesize a range of cyclic oligonucleotide and have recently emerged as important regulators of phage infections6--8. Here, we identify two novel cGAS-like receptors (cGLRs) in the insect Drosophila melanogaster. We show that cGLR1 and cGLR2 activate Sting and NF-$kappa$B dependent antiviral immunity in response to infection with RNA or DNA viruses. cGLR1 is activated by dsRNA to produce the novel CDN 3'2'-cGAMP whereas cGLR2 produces a combination of 2'3'-cGAMP and 3'2' cGAMP in response to a yet unidentified stimulus. Our data establish cGAS as the founding member of a family of receptors sensing different types of nucleic acids and triggering immunity through production of CDNs beyond 2'3'-cGAMP.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
M. Krivega, C.M. Stiefel, S. Karbassi, L.L. Andersen, N.K. Chunduri, N. Donnelly, A. Pichlmair, Z. Storchová
Genotoxic Stress in Constitutive Trisomies Induces Autophagy and the Innate Immune Response via the cGAS-STING Pathway Journal Article
In: Communications Biology, vol. 4, no. 1, pp. 831, 2021, ISSN: 2399-3642.
@article{Krivega2021,
title = {Genotoxic Stress in Constitutive Trisomies Induces Autophagy and the Innate Immune Response via the cGAS-STING Pathway},
author = {M. Krivega and C.M. Stiefel and S. Karbassi and L.L. Andersen and N.K. Chunduri and N. Donnelly and A. Pichlmair and Z. Storchová},
url = {https://doi.org/10.1038/s42003-021-02278-9},
doi = {10.1038/s42003-021-02278-9},
issn = {2399-3642},
year = {2021},
date = {2021-07-02},
journal = {Communications Biology},
volume = {4},
number = {1},
pages = {831},
abstract = {Gain of even a single chromosome leads to changes in human cell physiology and uniform perturbations of specific cellular processes, including downregulation of DNA replication pathway, upregulation of autophagy and lysosomal degradation, and constitutive activation of the type I interferon response. Little is known about the molecular mechanisms underlying these changes. We show that the constitutive nuclear localization of TFEB, a transcription factor that activates the expression of autophagy and lysosomal genes, is characteristic of human trisomic cells. Constitutive nuclear localization of TFEB in trisomic cells is independent of mTORC1 signaling, but depends on the cGAS-STING activation. Trisomic cells accumulate cytoplasmic dsDNA, which activates the cGAS-STING signaling cascade, thereby triggering nuclear accumulation of the transcription factor IRF3 and, consequently, upregulation of interferon-stimulated genes. cGAS depletion interferes with TFEB-dependent upregulation of autophagy in model trisomic cells. Importantly, activation of both the innate immune response and autophagy occurs also in primary trisomic embryonic fibroblasts, independent of the identity of the additional chromosome. Our research identifies the cGAS-STING pathway as an upstream regulator responsible for activation of autophagy and inflammatory response in human cells with extra chromosomes, such as in Down syndrome or other aneuploidy-associated pathologies.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
A. Stukalov, V. Girault, V. Grass, O. Karayel, V. Bergant, C. Urban, D.A. Haas, Y. Huang, L. Oubraham, A. Wang, M.S. Hamad, A. Piras, F.M. Hansen, M.C. Tanzer, I. Paron, L. Zinzula, T. Enghleitner, M. Reinecke, T.M. Lavacca, R. Ehmann, R. Wölfel, J. Jores, B. Kuster, U. Protzer, R. Rad, J. Ziebuhr, V. Thiel, P. Scaturro, M. Mann, A. Pichlmair
Multilevel Proteomics Reveals Host Perturbations by SARS-CoV-2 and SARS-CoV Journal Article
In: Nature, 2021, ISSN: 0028-0836.
@article{Stukalov2021,
title = {Multilevel Proteomics Reveals Host Perturbations by SARS-CoV-2 and SARS-CoV},
author = {A. Stukalov and V. Girault and V. Grass and O. Karayel and V. Bergant and C. Urban and D.A. Haas and Y. Huang and L. Oubraham and A. Wang and M.S. Hamad and A. Piras and F.M. Hansen and M.C. Tanzer and I. Paron and L. Zinzula and T. Enghleitner and M. Reinecke and T.M. Lavacca and R. Ehmann and R. Wölfel and J. Jores and B. Kuster and U. Protzer and R. Rad and J. Ziebuhr and V. Thiel and P. Scaturro and M. Mann and A. Pichlmair},
doi = {10.1038/s41586-021-03493-4},
issn = {0028-0836},
year = {2021},
date = {2021-04-12},
journal = {Nature},
abstract = {The global emergence of SARS-CoV-2 urgently requires an in-depth understanding of molecular functions of viral proteins and their interactions with the host proteome. Several individual omics studies have extended our knowledge of COVID-19 pathophysiology1–10. Integration of such datasets to obtain a holistic view of virus-host interactions and to define the pathogenic properties of SARS-CoV-2 is limited by the heterogeneity of the experimental systems. We therefore conducted a concurrent multi-omics study of SARS-CoV-2 and SARS-CoV. Using state-of-the-art proteomics, we profiled the interactome of both viruses, as well as their influence on transcriptome, proteome, ubiquitinome and phosphoproteome in a lung-derived human cell line. Projecting these data onto the global network of cellular interactions revealed crosstalk between the perturbations taking place upon SARS-CoV-2 and SARS-CoV infections at different layers and identified unique and common molecular mechanisms of these closely related coronaviruses. The TGF-β pathway, known for its involvement in tissue fibrosis, was specifically dysregulated by SARS-CoV-2 ORF8 and autophagy by SARS-CoV-2 ORF3. The extensive dataset (available at https://covinet.innatelab.org) highlights many hotspots that can be targeted by existing drugs and it can guide rational design of virus- and host-directed therapies, which we exemplify by identifying kinase and MMPs inhibitors with potent antiviral effects against SARS-CoV-2.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
D. Lainšček, T. Fink, V. Forstnerič, I. Hafner-Bratkovič, S. Orehek, Ž. Strmšek, M. Manček-Keber, P. Pečan, H. Esih, Š. Malenšek, J. Aupič, P. Dekleva, T. Plaper, S. Vidmar, L. Kadunc, M. Benčina, N. Omersa, G. Anderluh, F. Pojer, K. Lau, D. Hacker, B.E. Correia, D. Peterhoff, R. Wagner, V. Bergant, A. Herrmann, A. Pichlmair, R. Jerala
A Nanoscaffolded Spike-RBD Vaccine Provides Protection against SARS-CoV-2 with Minimal Anti-Scaffold Response Journal Article
In: Vaccines, vol. 9, no. 5, 2021, ISSN: 2076-393X.
@article{vaccines9050431,
title = {A Nanoscaffolded Spike-RBD Vaccine Provides Protection against SARS-CoV-2 with Minimal Anti-Scaffold Response},
author = {D. Lainšček and T. Fink and V. Forstnerič and I. Hafner-Bratkovič and S. Orehek and Ž. Strmšek and M. Manček-Keber and P. Pečan and H. Esih and Š. Malenšek and J. Aupič and P. Dekleva and T. Plaper and S. Vidmar and L. Kadunc and M. Benčina and N. Omersa and G. Anderluh and F. Pojer and K. Lau and D. Hacker and B.E. Correia and D. Peterhoff and R. Wagner and V. Bergant and A. Herrmann and A. Pichlmair and R. Jerala},
url = {https://www.mdpi.com/2076-393X/9/5/431},
doi = {10.3390/vaccines9050431},
issn = {2076-393X},
year = {2021},
date = {2021-01-01},
journal = {Vaccines},
volume = {9},
number = {5},
abstract = {The response of the adaptive immune system is augmented by multimeric presentation of a specific antigen, resembling viral particles. Several vaccines have been designed based on natural or designed protein scaffolds, which exhibited a potent adaptive immune response to antigens; however, antibodies are also generated against the scaffold, which may impair subsequent vaccination. In order to compare polypeptide scaffolds of different size and oligomerization state with respect to their efficiency, including anti-scaffold immunity, we compared several strategies of presentation of the RBD domain of the SARS-CoV-2 spike protein, an antigen aiming to generate neutralizing antibodies. A comparison of several genetic fusions of RBD to different nanoscaffolding domains (foldon, ferritin, lumazine synthase, and β-annulus peptide) delivered as DNA plasmids demonstrated a strongly augmented immune response, with high titers of neutralizing antibodies and a robust T-cell response in mice. Antibody titers and virus neutralization were most potently enhanced by fusion to the small β-annulus peptide scaffold, which itself triggered a minimal response in contrast to larger scaffolds. The β-annulus fused RBD protein increased residence in lymph nodes and triggered the most potent viral neutralization in immunization by a recombinant protein. Results of the study support the use of a nanoscaffolding platform using the β-annulus peptide for vaccine design.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
A.C. Gonzalez-Perez, M. Stempel, E. Wyler, C. Urban, A. Piras, T. Hennig, S. Ganskih, Y. Wei, A. Heim, M. Landthaler, A. Pichlmair, L. Dölken, M. Munschauer, F. Erhard, M.M. Brinkmann, S.M. Horner
The Zinc Finger Antiviral Protein ZAP Restricts Human Cytomegalovirus and Selectively Binds and Destabilizes Viral UL4/UL5 Transcripts Journal Article
In: mBio, vol. 12, no. 3, pp. e02683-20, 2021.
@article{doi:10.1128/mBio.02683-20,
title = {The Zinc Finger Antiviral Protein ZAP Restricts Human Cytomegalovirus and Selectively Binds and Destabilizes Viral \textit{UL4}/\textit{UL5} Transcripts},
author = {A.C. Gonzalez-Perez and M. Stempel and E. Wyler and C. Urban and A. Piras and T. Hennig and S. Ganskih and Y. Wei and A. Heim and M. Landthaler and A. Pichlmair and L. Dölken and M. Munschauer and F. Erhard and M.M. Brinkmann and S.M. Horner},
doi = {10.1128/mBio.02683-20 URL = https://journals.asm.org/doi/abs/10.1128/mBio.02683-20},
year = {2021},
date = {2021-01-01},
journal = {mBio},
volume = {12},
number = {3},
pages = {e02683-20},
abstract = {Viral infections have a large impact on society, leading to major human and economic losses and even global instability. So far, many viral infections, including human cytomegalovirus (HCMV) infection, are treated with a small repertoire of drugs, often accompanied by the occurrence of resistant mutants. Interferon-stimulated gene products (ISGs) play a crucial role in early infection control. The ISG zinc finger CCCH-type antiviral protein 1 (ZAP/ZC3HAV1) antagonizes several RNA viruses by binding to CG-rich RNA sequences, whereas its effect on DNA viruses is less well understood. Here, we decipher the role of ZAP in the context of human cytomegalovirus (HCMV) infection, a β-herpesvirus that is associated with high morbidity in immunosuppressed individuals and newborns. We show that expression of the two major isoforms of ZAP, ZAP-S and ZAP-L, is induced during HCMV infection and that both negatively affect HCMV replication. Transcriptome and proteome analyses demonstrated that the expression of ZAP results in reduced viral mRNA and protein levels and decelerates the progression of HCMV infection. Metabolic RNA labeling combined with high-throughput sequencing (SLAM-seq) revealed that most of the gene expression changes late in infection result from the general attenuation of HCMV. Furthermore, at early stages of infection, ZAP restricts HCMV by destabilizing a distinct subset of viral mRNAs, particularly those from the previously uncharacterized UL4-UL6 HCMV gene locus. Through enhanced cross-linking immunoprecipitation and sequencing analysis (eCLIP-seq), we identified the transcripts expressed from this HCMV locus as the direct targets of ZAP. Moreover, our data show that ZAP preferentially recognizes not only CG, but also other cytosine-rich sequences, thereby expanding its target specificity. In summary, this report is the first to reveal direct targets of ZAP during HCMV infection, which strongly indicates that transcripts from the UL4-UL6 locus may play an important role for HCMV replication. IMPORTANCE Viral infections have a large impact on society, leading to major human and economic losses and even global instability. So far, many viral infections, including human cytomegalovirus (HCMV) infection, are treated with a small repertoire of drugs, often accompanied by the occurrence of resistant mutants. There is no licensed HCMV vaccine in sight to protect those most at risk, particularly immunocompromised individuals or pregnant women who might otherwise transmit the virus to the fetus. Thus, the identification of novel intervention strategies is urgently required. In this study, we show that ZAP decelerates the viral gene expression cascade, presumably by selectively handpicking a distinct set of viral transcripts for degradation. Our study illustrates the potent role of ZAP as an HCMV restriction factor and sheds light on a possible role for UL4 and/or UL5 early during infection, paving a new avenue for the exploration of potential targets for novel therapies.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
D. Stadler, M. Kächele, A.N. Jones, J. Hess, C. Urban, J. Schneider, Y. Xia, A. Oswald, F. Nebioglu, R. Bester, F. Lasitschka, M. Ringelhan, C. Ko, W. Chou, A. Geerlof, M.A. van de Klundert, J.M. Wettengel, P. Schirmacher, M. Heikenwälder, S. Schreiner, R. Bartenschlager, A. Pichlmair, M. Sattler, K. Unger, U. Protzer
Interferon-induced Degradation of the Persistent Hepatitis B Virus cccDNA Form Depends on ISG20 Journal Article
In: EMBO reports, vol. 22, no. 6, pp. e49568, 2021.
@article{https://doi.org/10.15252/embr.201949568,
title = {Interferon-induced Degradation of the Persistent Hepatitis B Virus cccDNA Form Depends on ISG20},
author = {D. Stadler and M. Kächele and A.N. Jones and J. Hess and C. Urban and J. Schneider and Y. Xia and A. Oswald and F. Nebioglu and R. Bester and F. Lasitschka and M. Ringelhan and C. Ko and W. Chou and A. Geerlof and M.A. van de Klundert and J.M. Wettengel and P. Schirmacher and M. Heikenwälder and S. Schreiner and R. Bartenschlager and A. Pichlmair and M. Sattler and K. Unger and U. Protzer},
url = {https://www.embopress.org/doi/abs/10.15252/embr.201949568},
doi = {https://doi.org/10.15252/embr.201949568},
year = {2021},
date = {2021-01-01},
journal = {EMBO reports},
volume = {22},
number = {6},
pages = {e49568},
abstract = {Abstract Hepatitis B virus (HBV) persists by depositing a covalently closed circular DNA (cccDNA) in the nucleus of infected cells that cannot be targeted by available antivirals. Interferons can diminish HBV cccDNA via APOBEC3-mediated deamination. Here, we show that overexpression of APOBEC3A alone is not sufficient to reduce HBV cccDNA that requires additional treatment of cells with interferon indicating involvement of an interferon-stimulated gene (ISG) in cccDNA degradation. Transcriptome analyses identify ISG20 as the only type I and II interferon-induced, nuclear protein with annotated nuclease activity. ISG20 localizes to nucleoli of interferon-stimulated hepatocytes and is enriched on deoxyuridine-containing single-stranded DNA that mimics transcriptionally active, APOBEC3A-deaminated HBV DNA. ISG20 expression is detected in human livers in acute, self-limiting but not in chronic hepatitis B. ISG20 depletion mitigates the interferon-induced loss of cccDNA, and co-expression with APOBEC3A is sufficient to diminish cccDNA. In conclusion, non-cytolytic HBV cccDNA decline requires the concerted action of a deaminase and a nuclease. Our findings highlight that ISGs may cooperate in their antiviral activity that may be explored for therapeutic targeting.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
A.B.C. Schuren, I.G.J. Boer, E.M. Bouma, M.L. Van de Weijer, A.I. Costa, P. Hubel, A. Pichlmair, R.J. Lebbink, E.J.H.J. Wiertz
The UFM1 Pathway Impacts HCMV US2-Mediated Degradation of HLA Class I Journal Article
In: Molecules, vol. 26, no. 2, 2021, ISSN: 1420-3049.
@article{molecules26020287,
title = {The UFM1 Pathway Impacts HCMV US2-Mediated Degradation of HLA Class I},
author = {A.B.C. Schuren and I.G.J. Boer and E.M. Bouma and M.L. Van de Weijer and A.I. Costa and P. Hubel and A. Pichlmair and R.J. Lebbink and E.J.H.J. Wiertz},
url = {https://www.mdpi.com/1420-3049/26/2/287},
doi = {10.3390/molecules26020287},
issn = {1420-3049},
year = {2021},
date = {2021-01-01},
journal = {Molecules},
volume = {26},
number = {2},
abstract = {To prevent accumulation of misfolded proteins in the endoplasmic reticulum, chaperones perform quality control on newly translated proteins and redirect misfolded proteins to the cytosol for degradation by the ubiquitin-proteasome system. This pathway is called ER-associated protein degradation (ERAD). The human cytomegalovirus protein US2 induces accelerated ERAD of HLA class I molecules to prevent immune recognition of infected cells by CD8+ T cells. Using US2-mediated HLA-I degradation as a model for ERAD, we performed a genome-wide CRISPR/Cas9 library screen to identify novel cellular factors associated with ERAD. Besides the identification of known players such as TRC8, p97, and UBE2G2, the ubiquitin-fold modifier1 (UFM1) pathway was found to affect degradation of HLA-I. UFMylation is a post-translational modification resembling ubiquitination. Whereas we observe ubiquitination of HLA-I, no UFMylation was detected on HLA-I or several other proteins involved in degradation of HLA-I, suggesting that the UFM1 pathway impacts ERAD in a different manner than ubiquitin. Interference with the UFM1 pathway seems to specifically inhibit the ER-to-cytosol dislocation of HLA-I. In the absence of detectable UFMylation of HLA-I, UFM1 may contribute to US2-mediated HLA-I degradation by misdirecting protein sorting indirectly. Mass spectrometry analysis of US2-expressing cells showed that ribosomal proteins are a major class of proteins undergoing extensive UFMylation; the role of these changes in protein degradation may be indirect and remains to be established.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Prof. Dr. Andreas Pichlmair
Immunopathology of Virus Infections Laboratory, Institute of Virology, Technical University of Munich
Einsteinstr. 25, D-81675 Munich, Germany
tel: +49 (0) 89 4140 9270, email: andreas.pichlmair@tum.de
https://www.mri.tum.de/datenschutz
Der Datenschutzbeauftragte des Klinikums ist unter Klinikum rechts der Isar, Stabsstelle Datenschutz, 81664 München oder unter 089/4140-0 oder datenschutz@mri.tum.de zu erreichen.
Der Datenschutzbeauftragte der TU München ist unter beauftragter@datenschutz.tum.de zu erreichen.