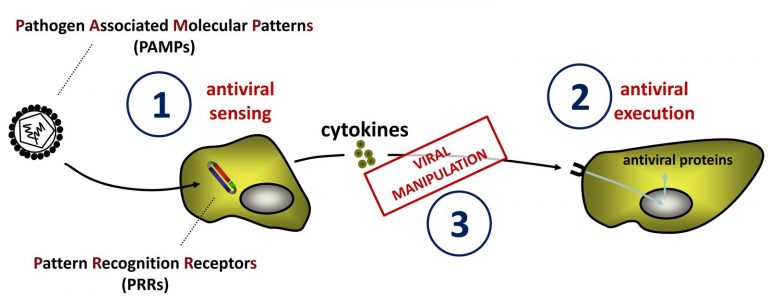
Our lab is interested in the interaction of viral structures (proteins and nucleic acids) with host factors and the relevance for antiviral immunity. We aim to gain functional and mechanistic insights in the interplay between viruses and the organism by studying virus-host interactions and protein expression profiles that are elicited by viral infections. Through this approach we identify yet unstudied proteins and pathways that we are further testing in focused hypothesis-driven approaches that include testing of interactions on molecular basis, in vitro cell culture assays and in vivomodels using genetically modified animals.
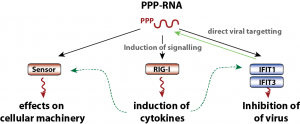
Though comprehensive knowledge on changes in the transcriptome, comparable little is known on the global changes of the proteome after infection with individual viruses. We are assessing virally-induced changes in the global composition of the proteome as well as specific post-translational modifications.
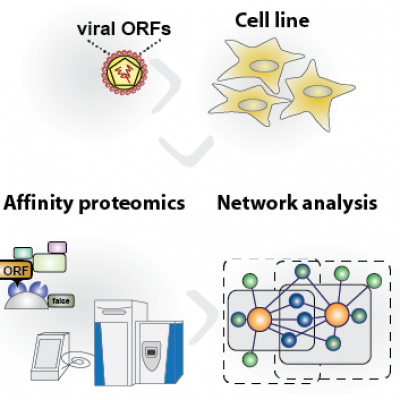
Viruses require the host cellular machinery to replicate. We use viruses go guide us to cellular proteins and pathways that are determining virus pathogenicity. To this aim we are using mass spectrometry to study cellular binding partners of viral proteins using systems biology approaches (Pichlmair et al., Nature 2012). We identified 600 cellular proteins that are binding to of viral immune modulators (iVIMs). We are now complementing this survey to assess the functional consequences for antiviral immunity. This survey so led to identification of novel modes of transcriptional regulation by an orthomyxovirus that specifically affects genes required for host defense (Haas et al., Plos Pathogens, 2018). Furthermore, we identified a novel cell death pathway named Oxeiptosis that is targeted by viral proteins derived from diverse viruses. Intracellular reactive oxygen species (ROS) that are commonly generated during virus infections, engagement of toxic substances or teratogenic transformation of cells (Holze et al., Nature Immunology, 2018). The cellular protein KEAP1 senses increased ROS levels and activates a cell death cascades that involves the mitochondrial enzyme PGAM5 that dephosphorylates the protein AIFM1. This leads to cell death. Lack of oxeiptosis in mice induces hyperinflammation after virus infection, which is associated to increase immunopathology.
Our laboratory offers the opportunity to use top-notch technologies to study virus host interactions and that allow us high flexibility and independence:
We are running our own mass spectrometry platform (Q-Exactive plus HF mass analyzer coupled to a NanoLC 1200 UHPLC system), which gives us the opportunity to perform large scale screening experiments including testing protein-protein interactions, protein expression profiling, studying protein dynamics as well as post-translational modifications. Labmembers are preparing and analyzing their own samples after a short training period.
We are operating our own server (HPE ProLiant RackServer with 36 physical Cores/72 threats, 256 gByte RAM, 20TB HDD) and storage system (HPE StoreEasy, 70 TB), which we use for analysis ofmass spectrometry and other high content data. An own dedicated Bioinformatics subgroup is scripting all sort of algorithms that are essential to analyse mass spectrometry data and to generate high quality datasets.
The most recent addition to our laboratory. This system allows phenotypic screens and bridges the gap between mass spectrometry analysis (mostly discovering hundreds of candidates) to detailed functional and mechanistic studies on a very limited number of proteins. The incucyte S3 system allows fluorescence based screens of up to 584 conditions in a time-resolved manner. Combination with CRISPR/Cas9 and/or Drung screens will allow us to specifically perturb proteins identified by mass spectrometry analysis. A great tool that will transform the way we perform experiments in the lab.
Top modern FACS machine with a 96-well multisampler and the ability to test up to 13 colors in parallel.
A reliable and easy to use FPLC system with sample pump, the ability to mout 5 collumns in parallel, coupled to a fraction collector. This FPLC can be used to generate recombinant proteins, to fractionate cells or nuleic acids using various size exclusion or affinity collumns.
Prof. Dr. Andreas Pichlmair
Immunopathology of Virus Infections Laboratory, Institute of Virology, Technical University of Munich
Einsteinstr. 25, D-81675 Munich, Germany
tel: +49 (0) 89 4140 9270, email: andreas.pichlmair@tum.de
https://www.mri.tum.de/datenschutz
Der Datenschutzbeauftragte des Klinikums ist unter Klinikum rechts der Isar, Stabsstelle Datenschutz, 81664 München oder unter 089/4140-0 oder datenschutz@mri.tum.de zu erreichen.
Der Datenschutzbeauftragte der TU München ist unter beauftragter@datenschutz.tum.de zu erreichen.