Congratulations to Philipp Hubel and co-authors – our work on the innate immune network got published in Nature Immunology!
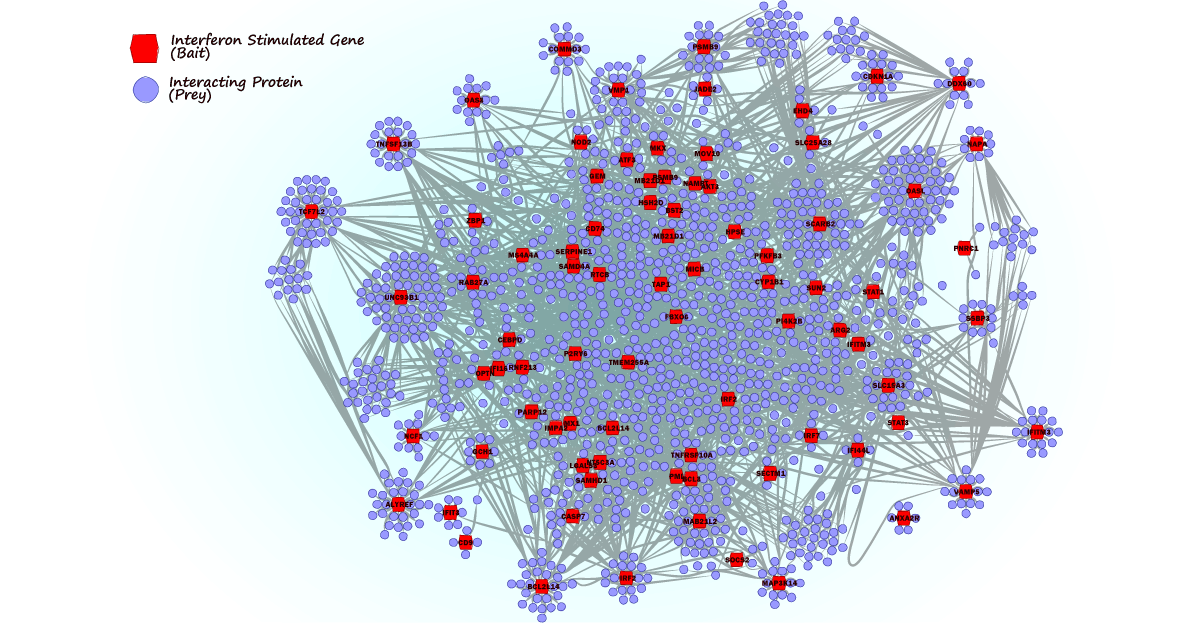
The body’s defense strategies against viral infections are as diverse as the attacks themselves. We conducted a survey to systematically investigate the interactions of interferon-stimulated genes (ISGs), which are at the center of coordinating the antiviral immune response. Through integrating data obtained by affinity purification followed by mass spectrometry (AP/MS), published datasets and functional validation experiments, we found many unknown interactions of ISGs, which sheds light on the overall organization of the innate immune system.
Have a look at the paper: “A protein-interaction network of interferon-stimulated genes extends the innate immune system landscape”