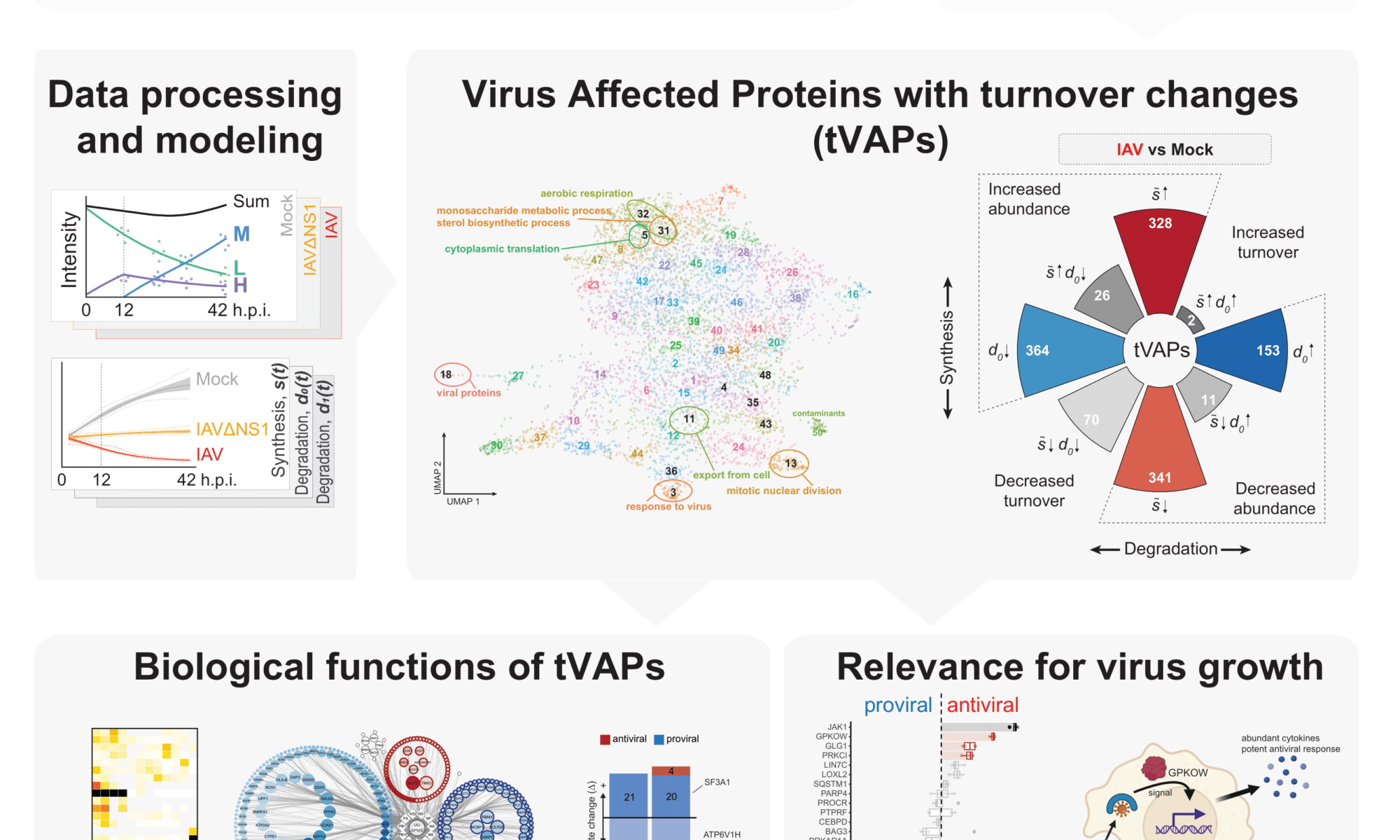
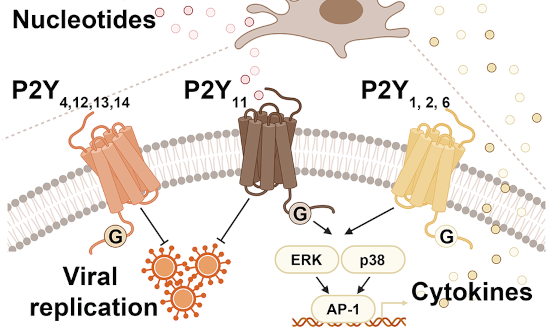
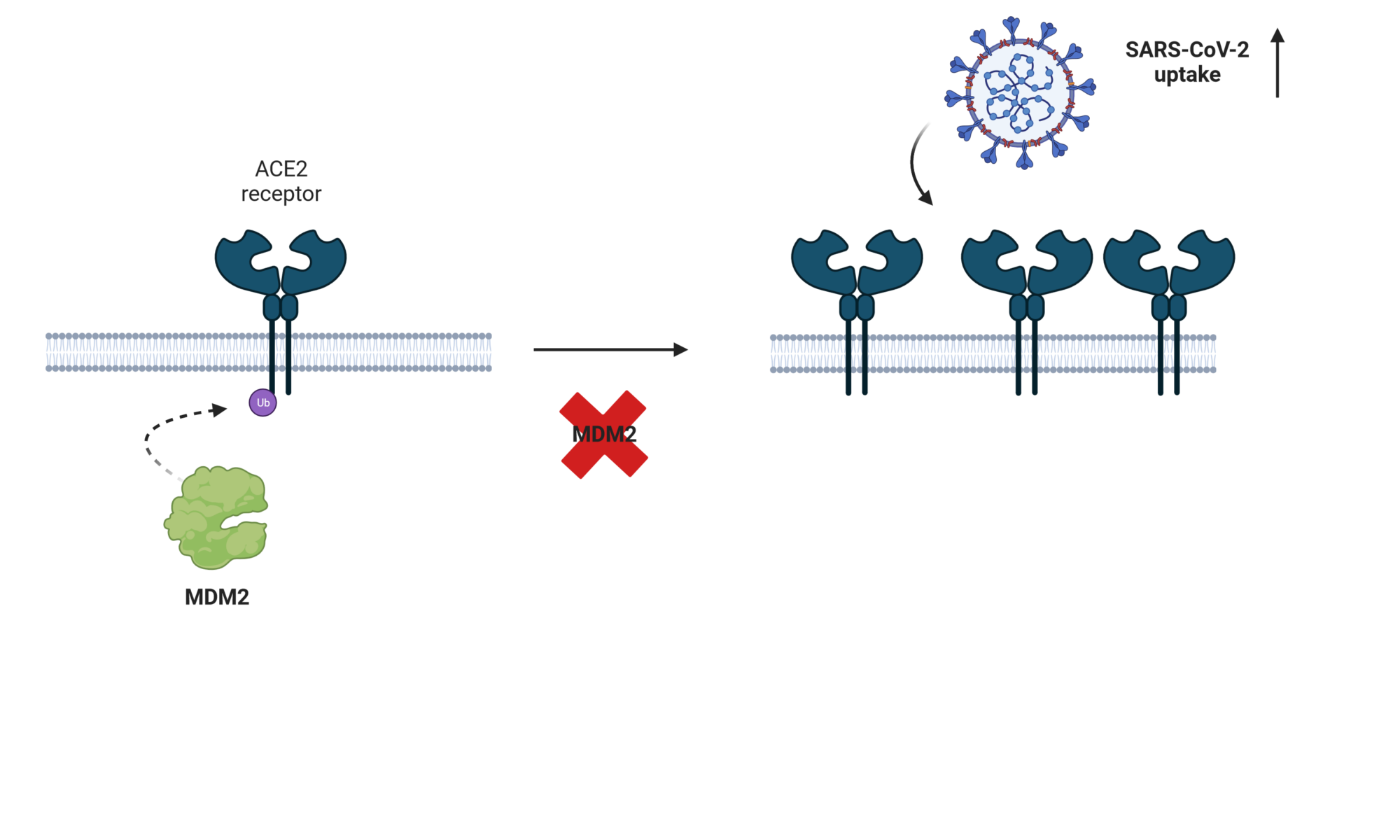
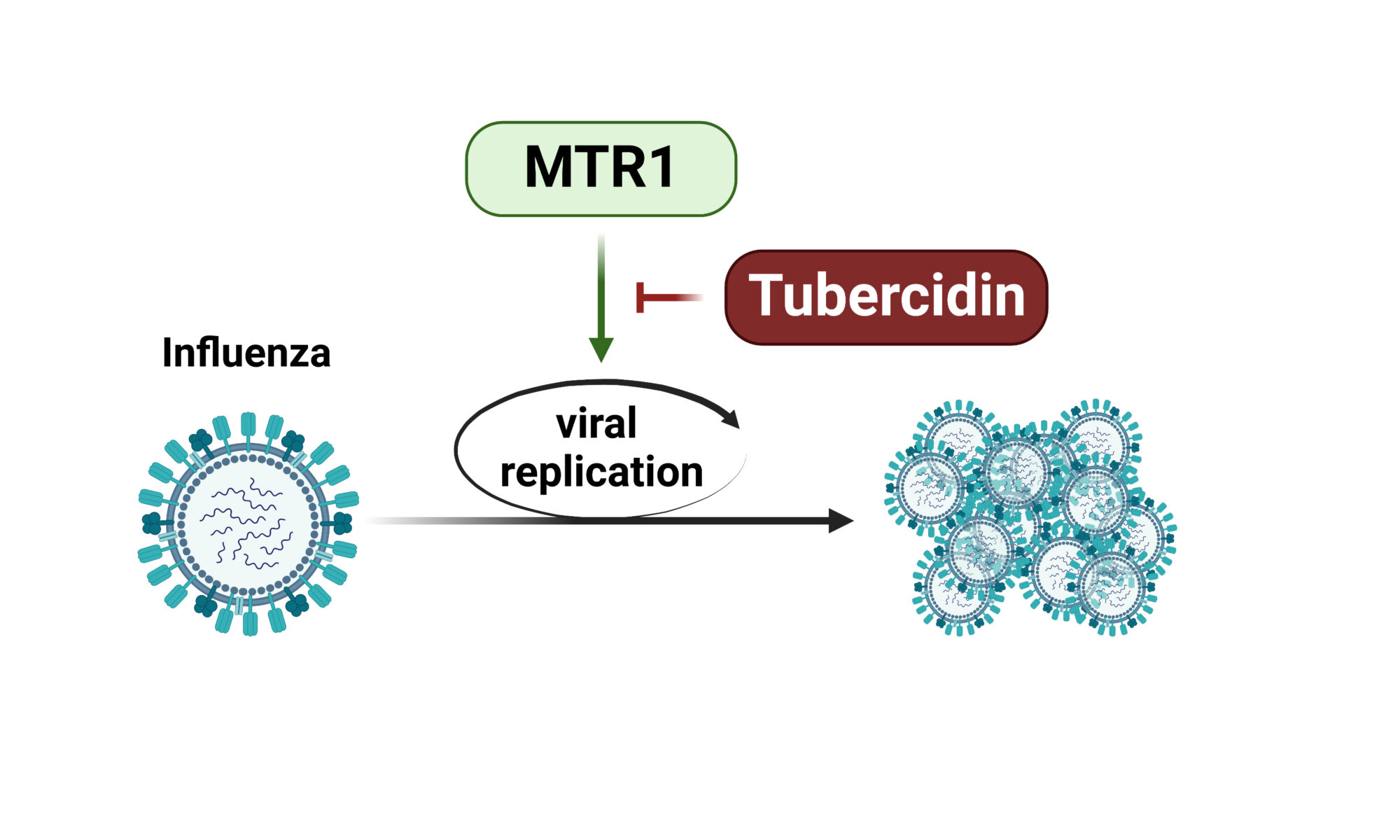
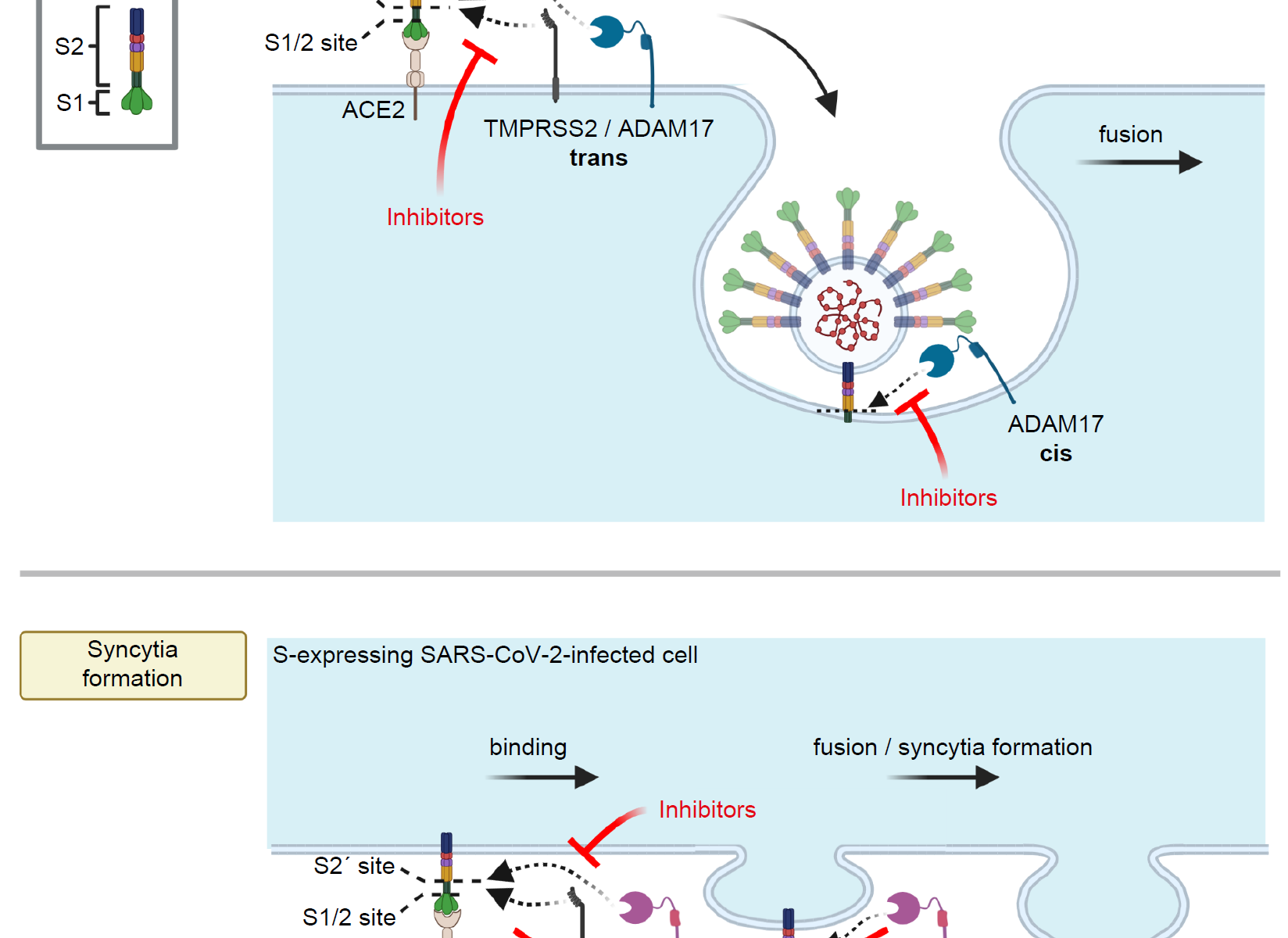
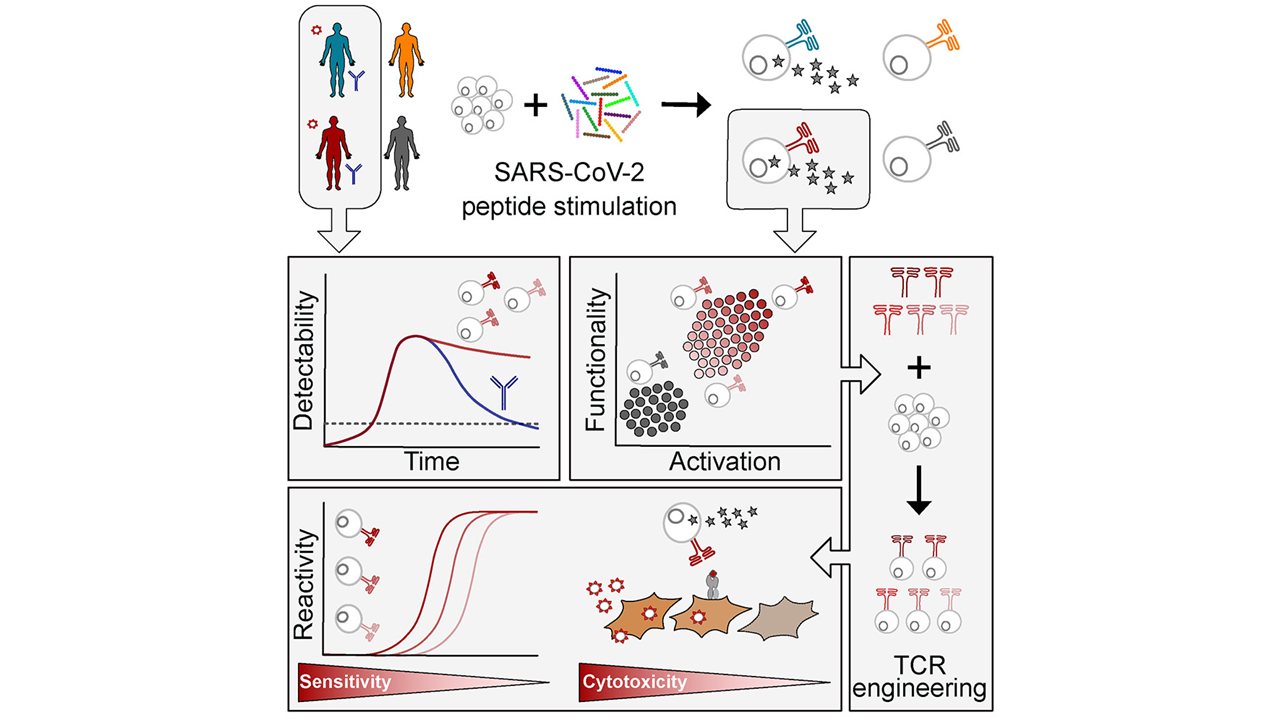
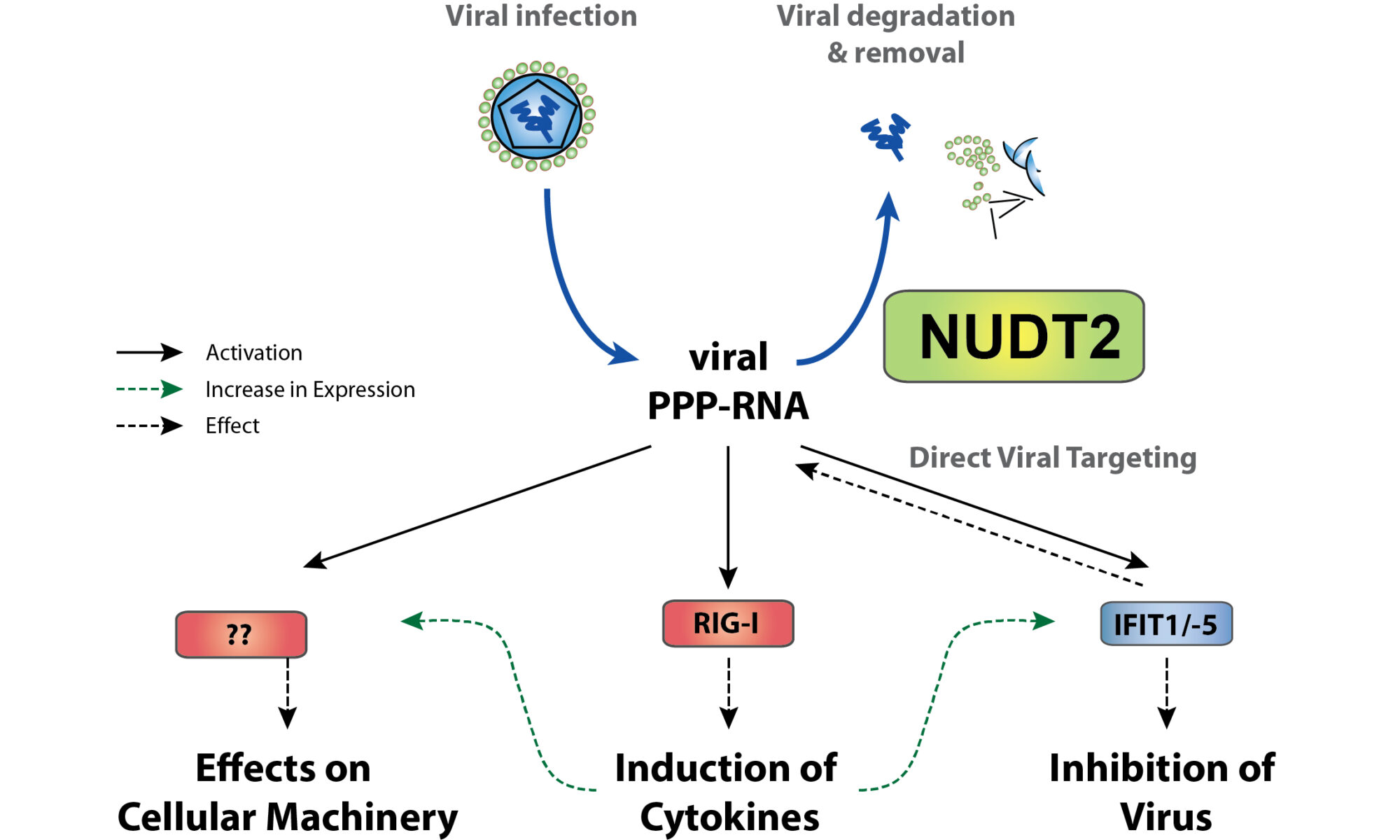
Protein turnover is crucial to the cellular response to internal and external changes. In our publication in Cell Systems, we systematically profiled the protein turnover changes in cells infected by the influenza A virus (IAV). We identified more than a thousand virus-affected proteins with turnover changes (tVAPs) and demonstrated their important roles in IAV infection by intersecting them with published IAV interactome and genome-wide screens and performing additional functional analyses. We further investigated a splicing factor GPKOW, which we found to be an antiviral tVAP that modulates the host’s innate immune response. The data can be explored here: https://pulsechase.innatelab.org/
Congratulations to Yiqi, Chris, Philipp and Alexey – it was a successful collaboration across multiple generations of Innates!
Read more here: Protein turnover regulation is critical for influenza A virus infection
Text by Yiqi.