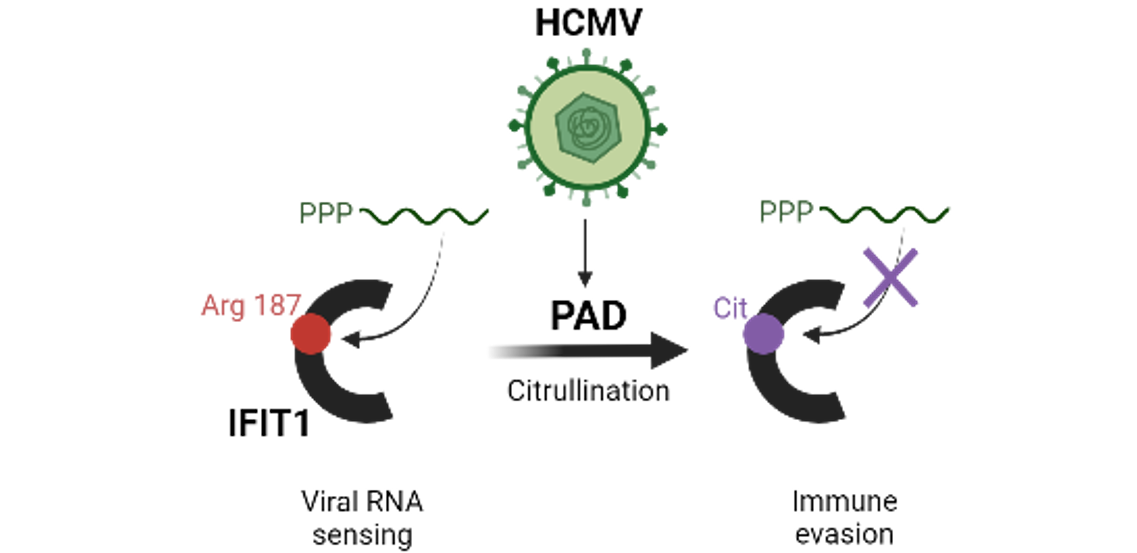
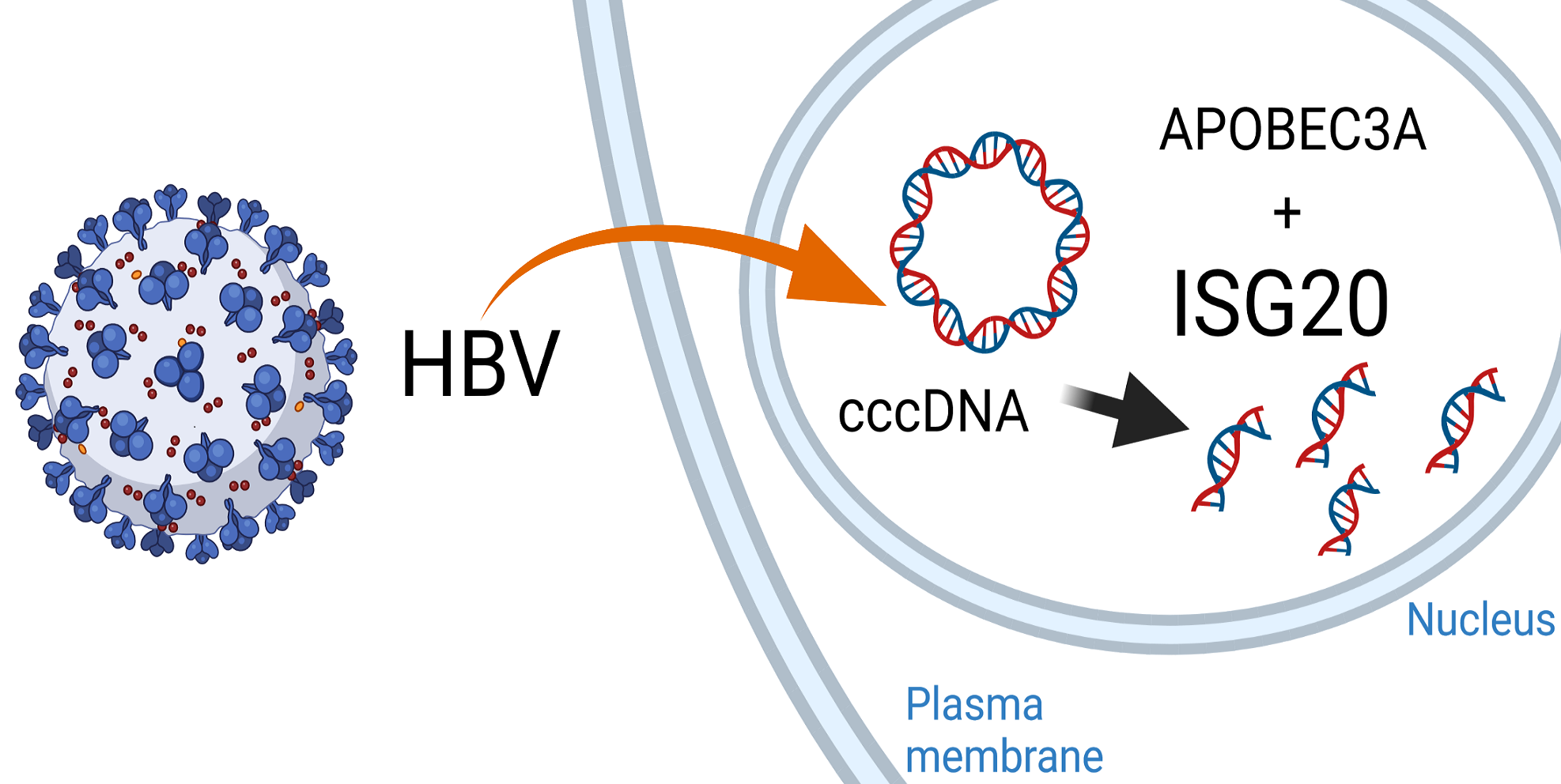
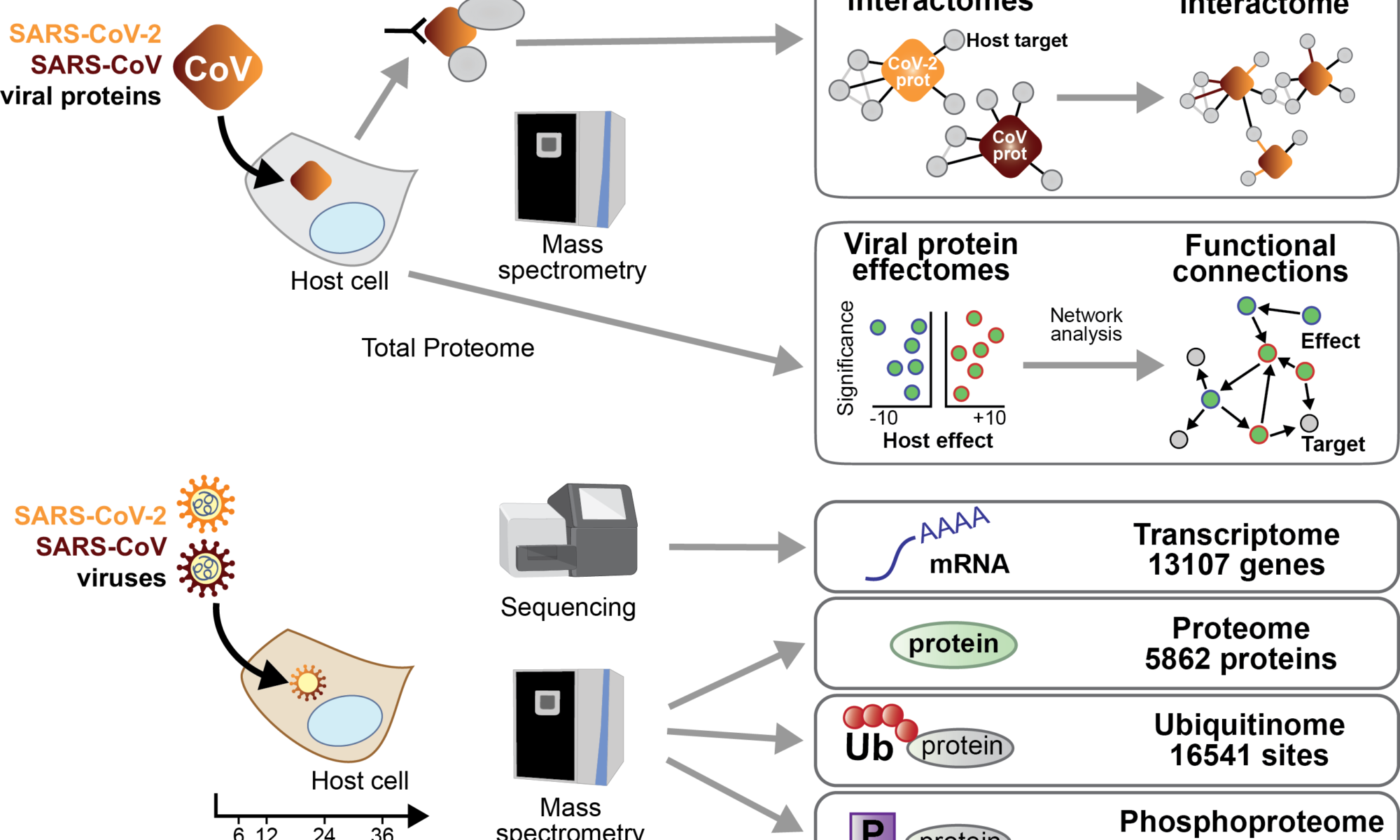
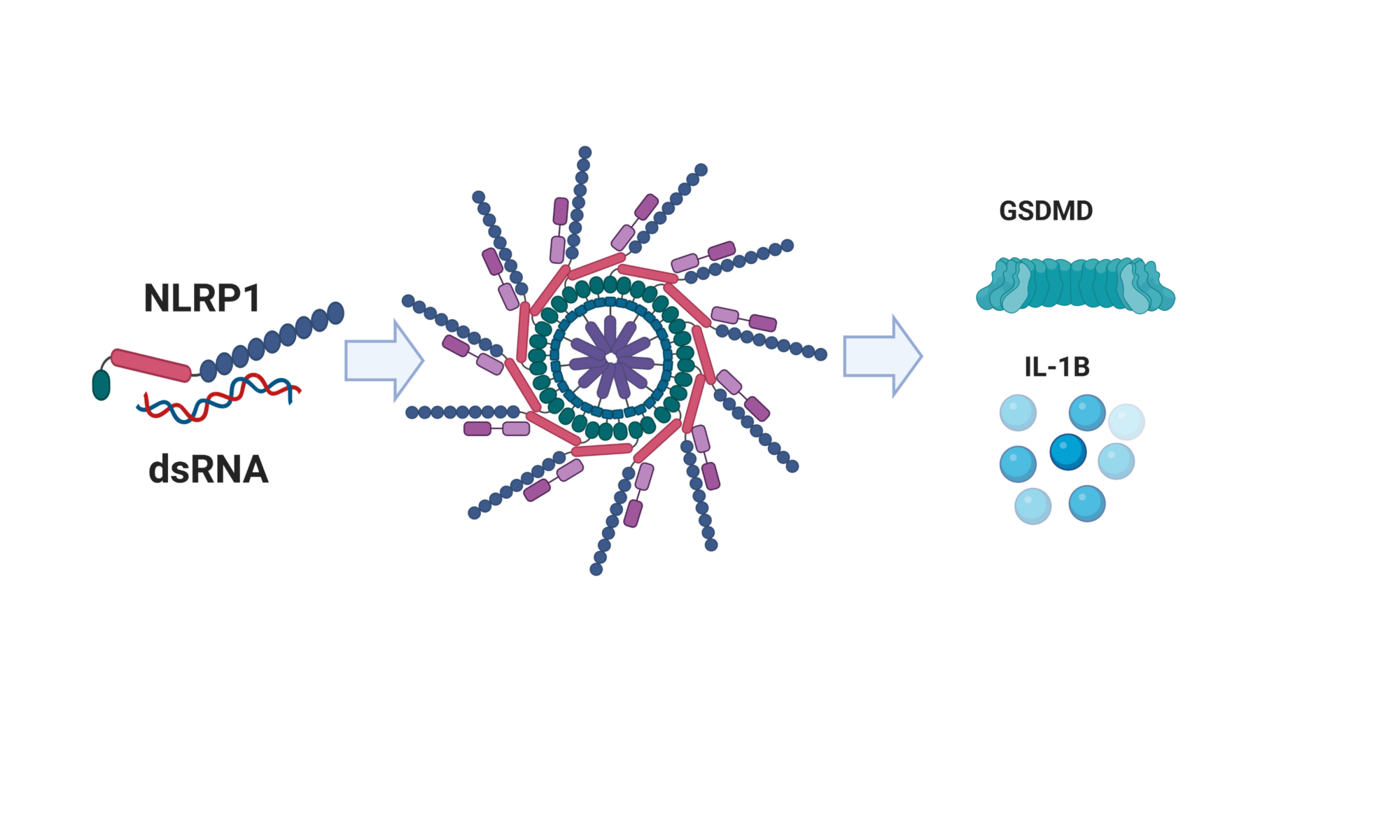
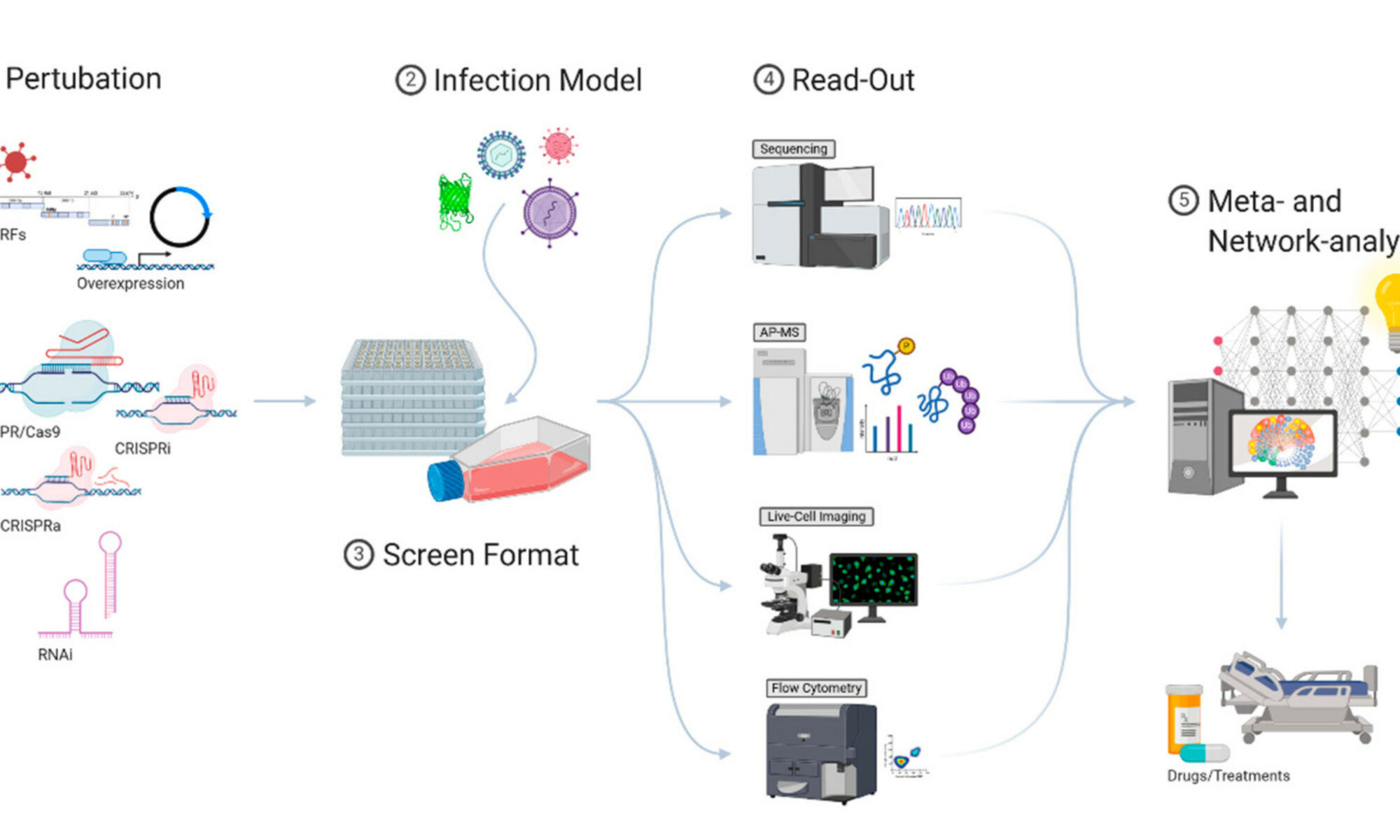
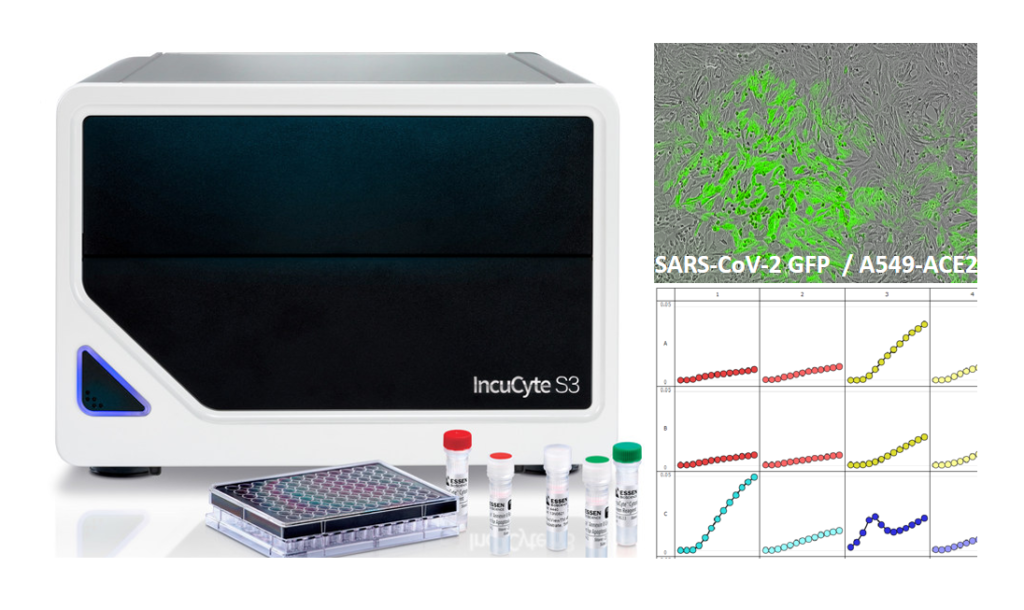
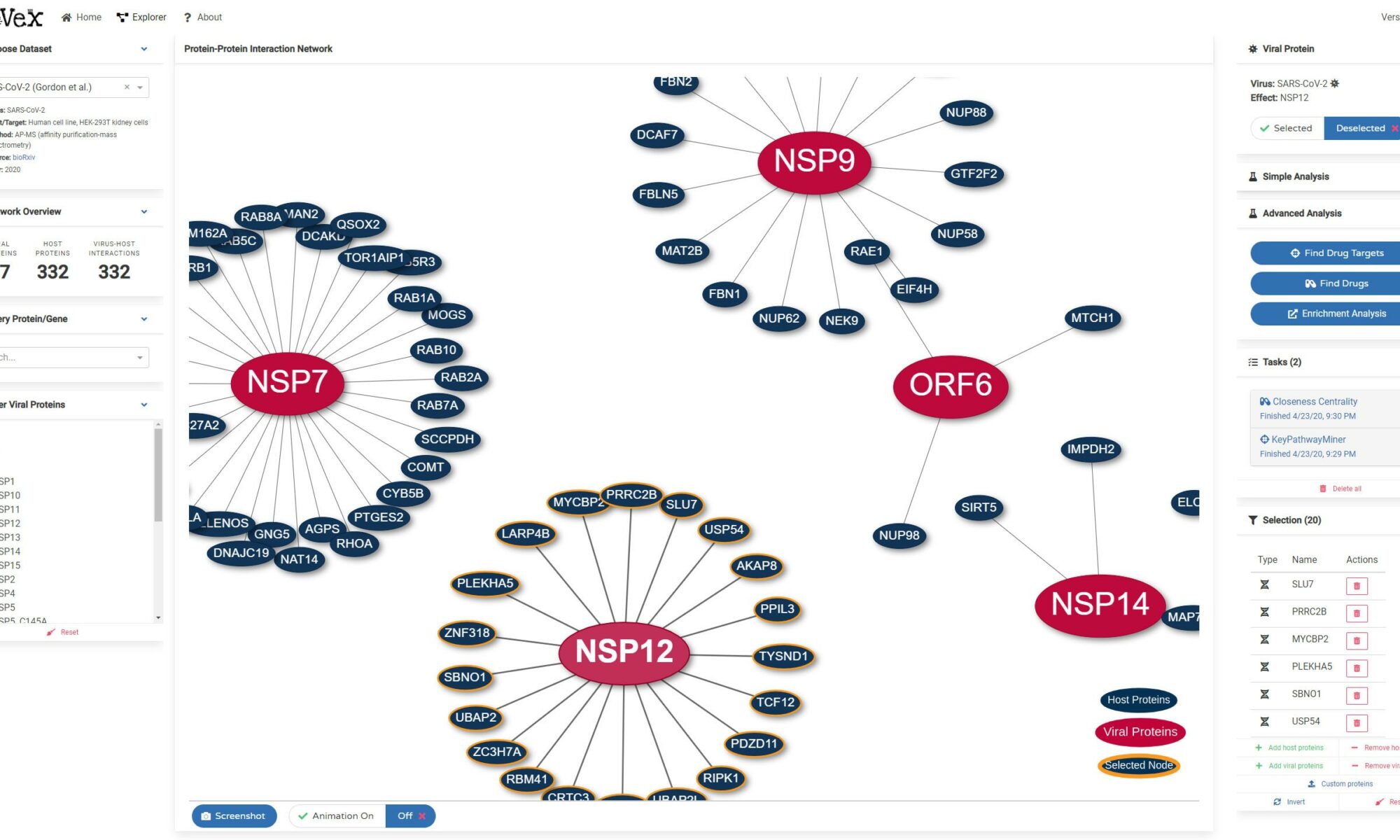
Gloria Griffante and colleagues (Santo Landolfo laboratory, University of Turin, Italy) discovered that HCMV infection induced protein citrullination, a post-translational modification catalyzed by PADs. In particular, the host defense protein IFIT1 was citrullinated by PAD2 and treatment with the enzyme impaired its ability to bind 5’PPP-RNA, thus constituting a novel immune evasion strategy. We contributed with mass-spectrometry and RNA-protein binding assays to characterize IFIT1 citrullination sites and PAD2-dependant modulation of its interaction with RNA.
Congratulations for this great work!
Read more in the Nature paper: Human cytomegalovirus-induced host protein citrullination is crucial for viral replication