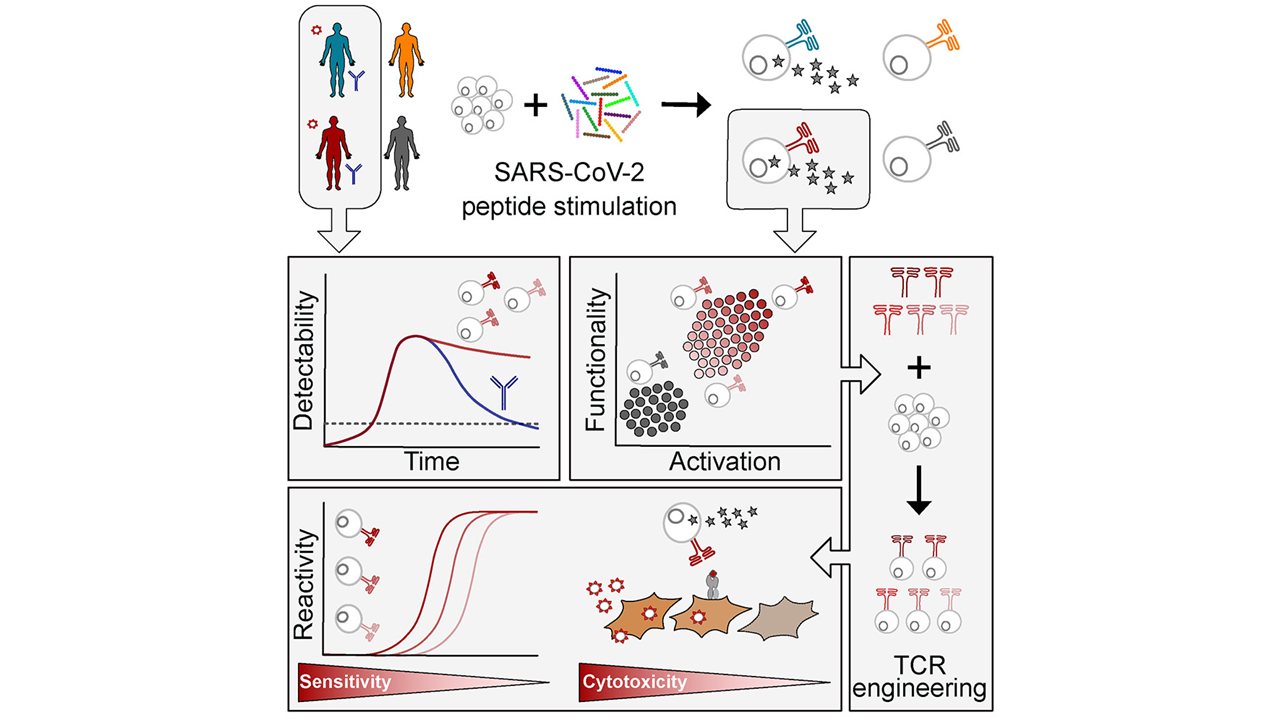
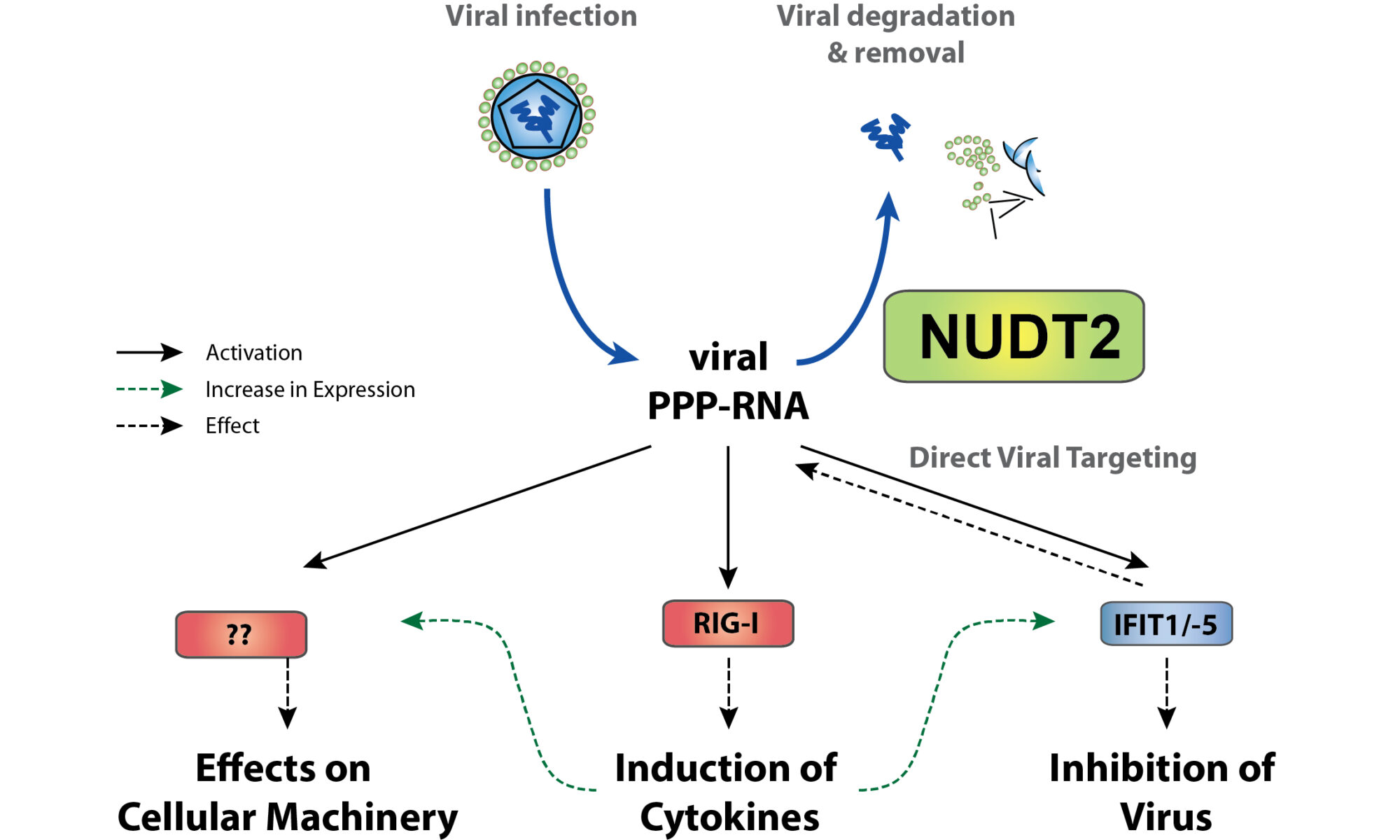
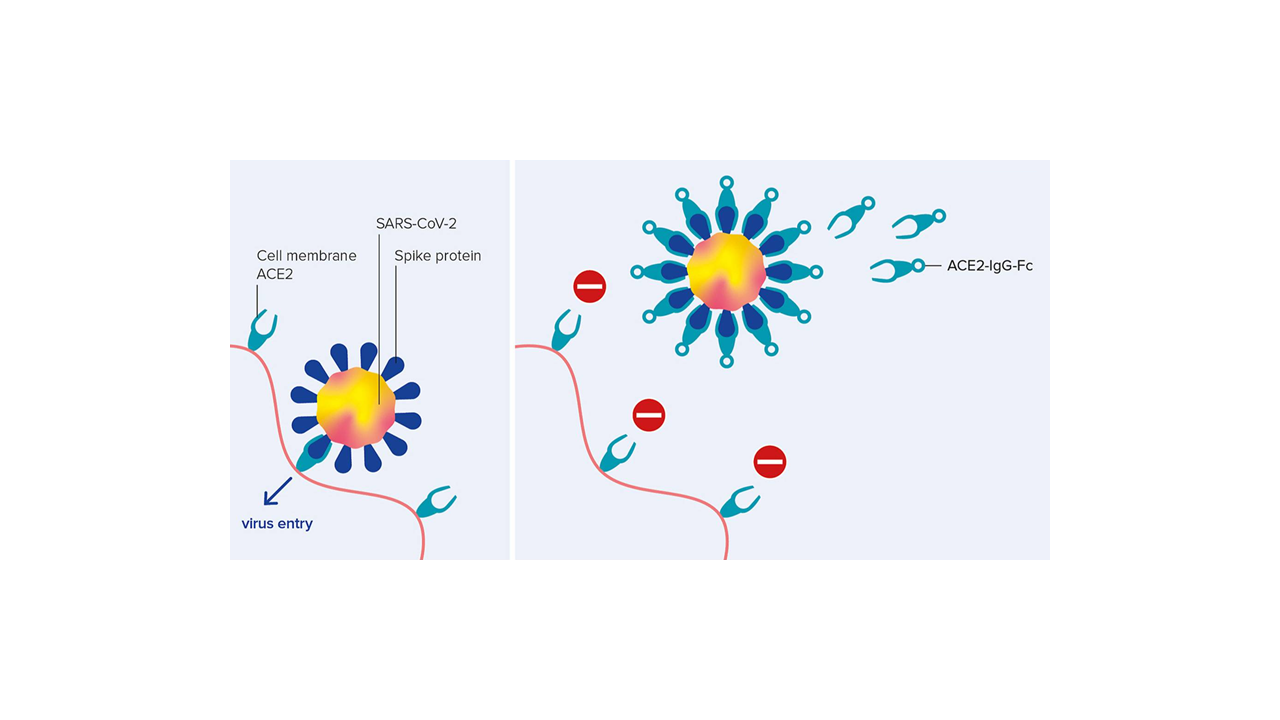
We are excited that our collaborative research center Transregio “Cell death decisions” (TRR353) has been approved for funding. TRR353 focuses on innovative and groundbreaking research related to the regulation of cell death decisions. Together with our colleagues at the University of Konstanz and the Albert Ludwig University of Freiburg, we will study how, why and when cells decide to die and how this can be exploited for therapeutic purposes. This funding will allow us to study the regulatory mechanisms of oxeiptosis, and the relationship of oxpeiptosis to other cell death pathways.
We are hiring PhD students for this project soon, so keep your eyes open!
Text by Lara.